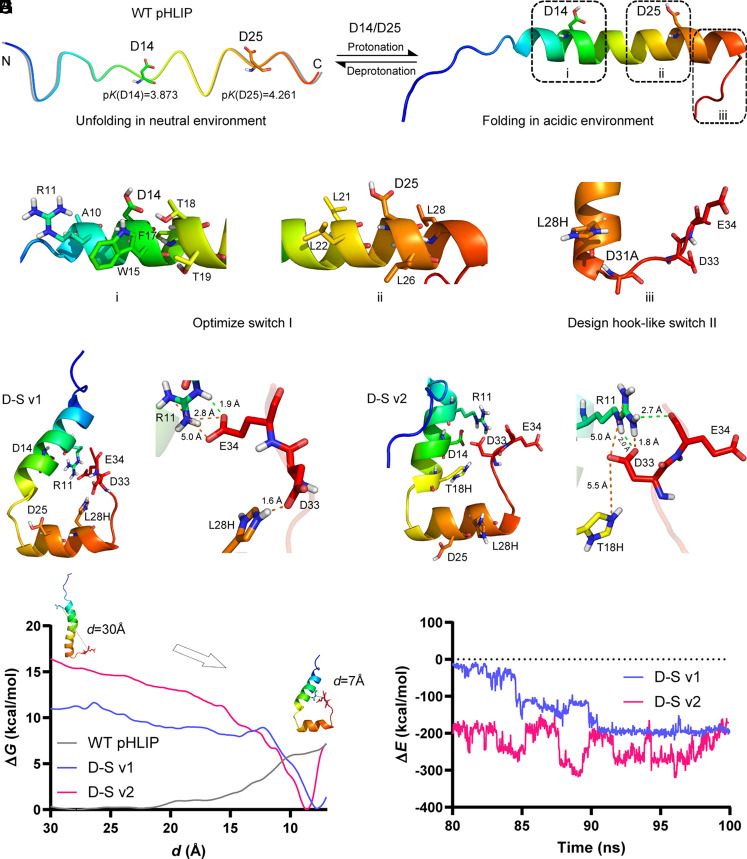
Fig. 1.
D-S pHLIP design. (A) The unfolding state of WT pHLIP in neutral environment. Two critical residues, D14 and D25 with their calculated pK values. (B) The folding state of WT pHLIP in acidic environment. Design regions for switch I (i and ii) and switch II (iii). (C and D) Residues that influence the pK values of D14 and D25. (E) A double mutation, L28H/D31A for the hook-like switch II. (F and G) Structural snapshots of the two D-S variants, i.e., L28H/D31A/L21F/L22G (D-S v1) and L28H/D31A/L21F/L22G/T18H (D-S v2) generated from the trajectories of their 100 ns MD simulations. Electrostatic and hydrogen bonding interactions are colored by orange and green dotted lines, respectively. (H and I) Calculations of free-energy and interaction-energy for WT pHLIP and two D-S variants. d denotes the center-of-mass distance between the two residues of D33 and E34 in the C-terminal and R11 nearby the N-terminal. In (A–G), nonpolar hydrogens are hidden. N and C in (A) denote the N- and C-terminal of the peptide, respectively, wherein the C-terminal will insert into the phospholipid bilayer structure after folding.