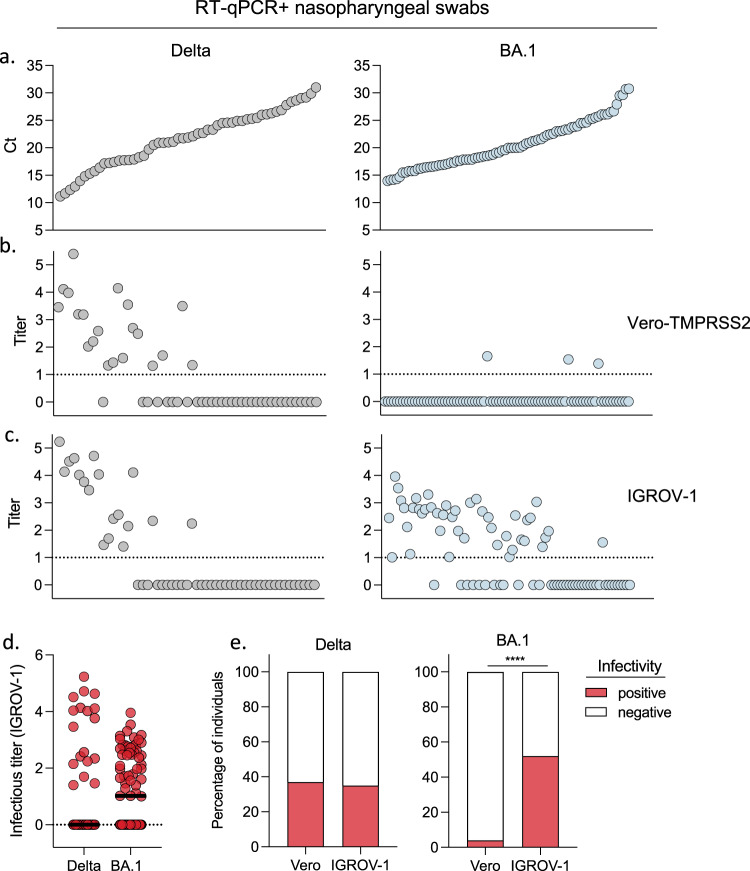
Fig. 1. Improved detection of infectious Omicron BA.1 in nasopharyngeal swabs using IGROV-1 cells.
A retrospective series of 135 RT+qPCR+ nasopharyngeal swabs from COVID-19 patients, harboring Delta (n = 53) or Omicron BA.1 (n = 82) variants was collected. a Viral RNA loads, measured by RT-qPCR. The samples were ranked from high to low viral RNA load (low to high Ct). Viral titers were measured in Vero-TMPRSS2 (b) and IGROV-1 cells (c). Delta and Omicron BA.1-positive samples are depicted in the left and right panels, respectively. d Comparison of infectious titers for Delta and BA.1 samples in IGROV-1 cells (left panel). e Percentage of samples harboring detectable infectious Delta (middle panel) or BA.1 virus (right panel) using Vero and IGROV-1 cells. A two-sided Chi-square test was performed ****p < 0.0001. Source data are provided as a Source Data file.