FIG 2.
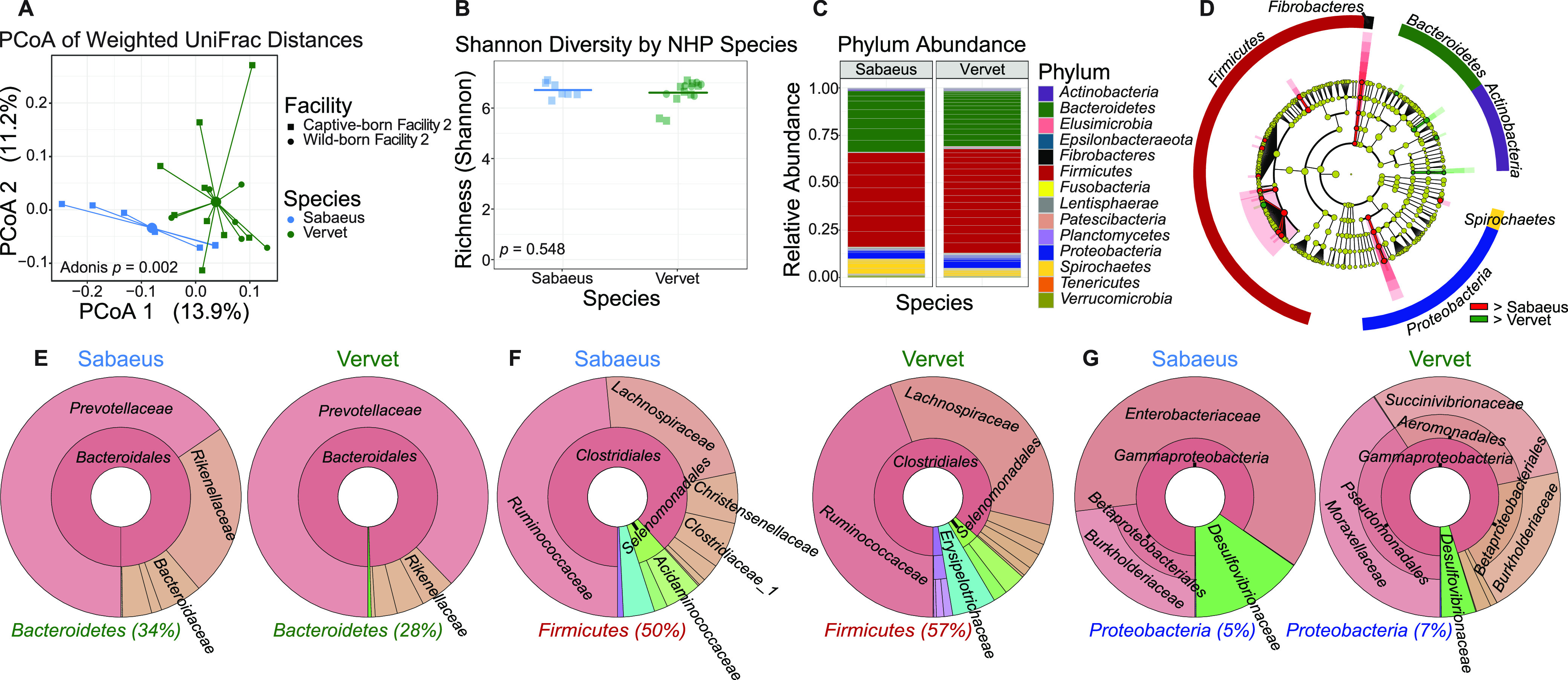
Gut microbiomes by Chlorocebus species differ significantly in β diversity. (A) Principal-coordinate analysis of weighted UniFrac distances (β diversity) of gut microbiota in sabaeus monkeys (n = 7) and vervets (n = 15). Significance between NHP species in fecal β diversity was assessed by Adonis. Lines represent the distance from each sample to the group’s centroid. (B) Shannon diversity (α diversity) comparison of fecal microbiomes between NHP species. Lines denote means. Significance between groups was determined by unpaired, two-way t test. (C) Relative abundance of bacterial families in NHP species measured by 16S rRNA gene sequencing. Color is by phylum, and line divisions are by family. (D) LEfSe cladogram representing all taxa in the fecal microbiome down to the genus level, with red (greater in sabaeus monkeys) or green (greater in vervets) nodes indicating significant differences. Gold nodes indicate no significant difference. Labels were restricted to the phylum level for ease of visualization. Full results of significant differences down to the genus level are available in Fig. S1, and all OTUs examined are available in Table S1. (E to G) Krona plots representing relative frequency of fecal Bacteroidetes (E), Firmicutes (F), and Proteobacteria (G) subtaxa comprising ≥5% phylum composition to the family level. Shown taxa are collapsed to the lowest common taxon. The percentage given after phyla in panels E to G is the percentage of total bacteria that phylum made up in the group.
