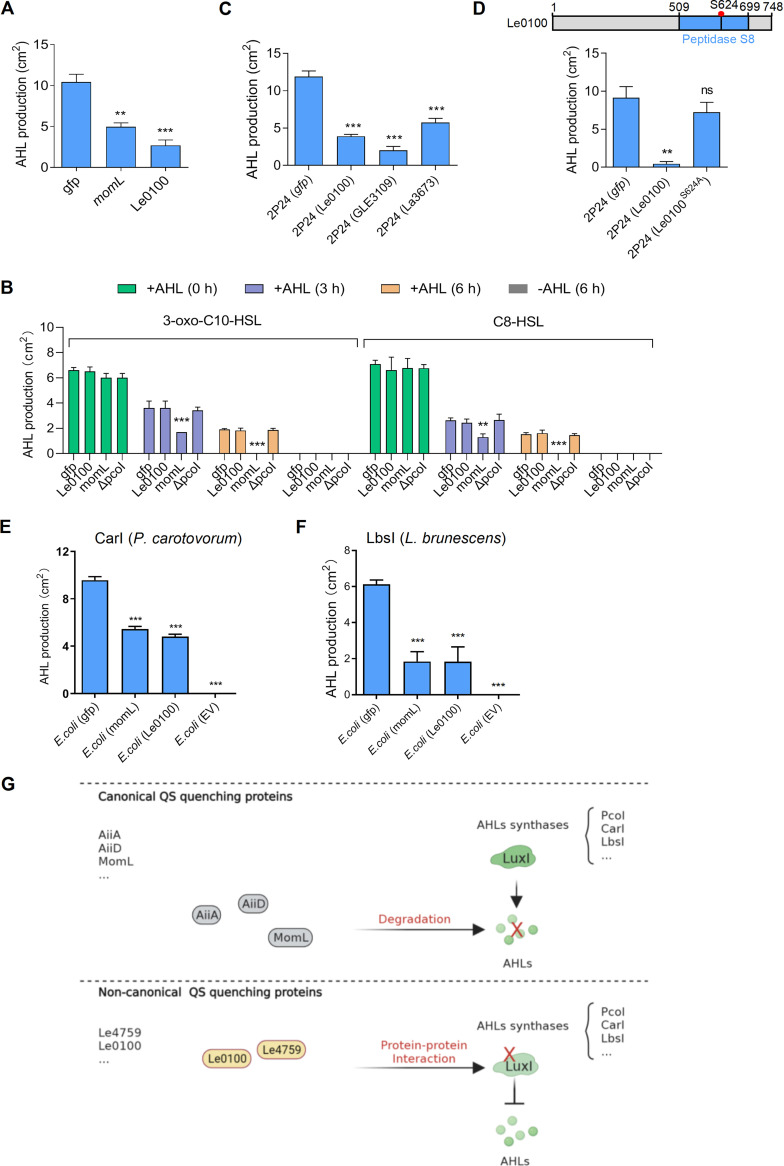
FIG 4.
Identification of Le0100 from Lysobacter enzymogenes, which encodes a predicated peptidase as an additional noncanonical AHL-quenching protein. (A) Heterogeneous expression of Le0100 in recombinant, pcoI-expressed E. coli significantly inhibited AHL production. (B) Expression of Le0100 could not degrade AHL. This assay was carried out similar to the assay of Fig. 2A. (C) Effects of heterogeneous expression of two Le0100 homologous genes on AHL production in pcoI-encoded Pseudomonas fluorescence 2P24 (2P24). GLE3109 and La3673 are Le0100 homologs from L. antibioticus OH13 and L. enzymogenes C3, respectively. (D) Identification of the key amino residue (S624) required for Le0100 to function as an AHL-quenching protein in 2P24. In panel C and D, all panels were the respective average data from three experiments. ± SD. ***, P < 0.0001; **, P < 0.01, assessed by one-way ANOVA. ns, no statistical significance. (E–F) Coexpression of Le0100 with carI (E) or lbsI (F) gene in E. coli significantly reduced AHL production. EV, empty vector. (G) A proposed model accounting for the different modes of action of those well-characterized canonical AHL-degrading enzymes and noncanonical AHL-quenching proteins presented in this study. Canonical AHL-quenching proteins of AiiA from Bacillus sp. 240B1, MomL from Muricauda olearia, and AiiD from Ralstonia sp. XJ12B could degrade AHLs produced by various AHL synthases listed in the right. To achieve this, these canonical AHL-quenching proteins do not need to interact with AHL synthases. In contrast, the noncanonical AHL quorum-quenching protein presented in this study could not degrade AHLs, whereas they, represented by Le4759, could bind diverse AHL synthases through direct protein-protein interactions, thereby blocking AHL production.