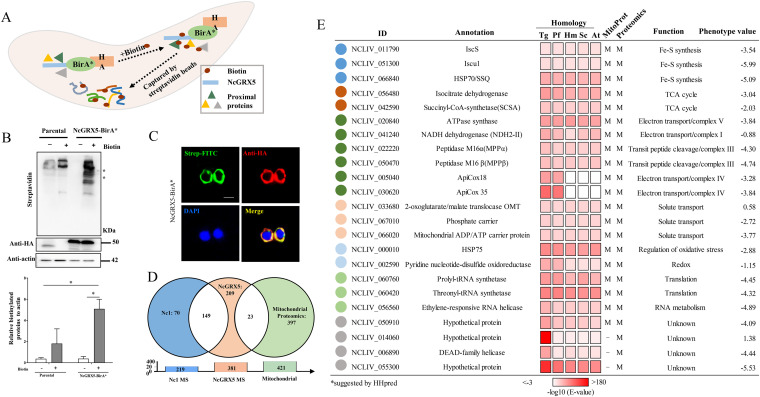
FIG 7.
Candidate NcGRX5-interacting proteins identified by the BioID method. (A) Diagram showing the BioID method. NcGRX5-BirA* is expressed in N. caninum. Biotinylated proteins were captured with streptavidin beads. (B) Western blot detection of biotinylated proteins in parental and NcGRX5-BirA*-expressing parasites ± biotin treatment. Lysates were probed with streptavidin-HRP, revealing an increase in biotinylated proteins in NcGRX5-BirA* parasites following biotin addition. The NcGRX5-BirA* fusion protein is expected to be ~50 kDa (arrowhead). Actin was used as a loading control. The biotinylated protein level normalized to β-actin was quantified by ImageJ from two independent experiments. Error bars represent standard errors. A one-way ANOVA was performed with Tukey’s correction for multiple comparisons *, P = 0.0452. (C) Immunofluorescence assay detection of biotinylated proteins in the NcGRX5-BirA* strain using streptavidin-FITC (green) and colocalization with NcGRX5. The location of NcGRX5 was detected by α-HA antibodies (red). Bar, 5 μm. (D) LC-MS was performed with 219 biotinylated proteins identified in parental parasites and 381 in NcGRX5-BirA*-expressing parasites. A total of 149 common nonspecific proteins were identified and removed. The remaining 232 proteins in NcGRX5-BirA*-expressing parasites correspond to the T. gondii mitochondrial proteome, with only 23 candidate proteins located in the mitochondria. (E) Table showing the 23 candidate biotinylated proteins located in the mitochondria (for more information, see Data set 3). The possible localization of proteins located in mitochondria (M) was evaluated using the T. gondii Mitochondrial Proteome Database and predicted using MitoProt (20–22). These proteins were annotated via ToxoDB searches or sequence homology alignment with T. gondii (Tg), P. falciparum (Pf), H. sapiens (Hs), S. cerevisiae (Sc), and A. thaliana (At). Homology alignment was performed by HHpred using a hidden Markov model-based search. The phenotype value was obtained from the corresponding T. gondii homologous protein in ToxoDB.