FIG 2.
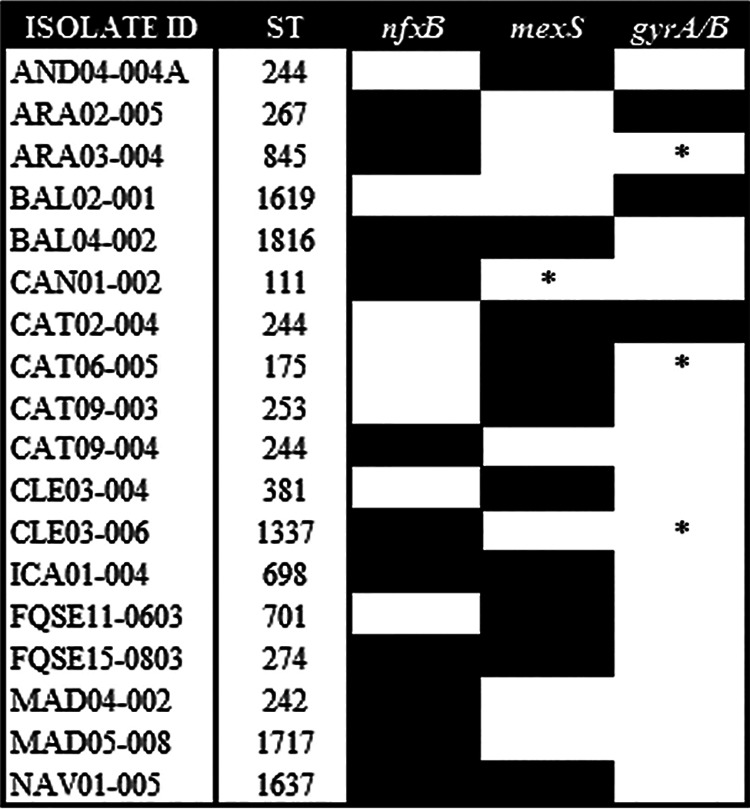
Diagram showing the genomic background dependence of the acquisition of ciprofloxacin resistance mutations leading to collateral sensitivity in clinical strains of P. aeruginosa. Classical mutations regularly implicated in ciprofloxacin resistance in clinical settings, within gyrAB, nfxB, and mexS genes, were acquired (black boxes) during ALE in the presence of ciprofloxacin for 3 days in populations belonging to 18 different clinical isolates. For gyrAB, we included only cases where genetic variations are present within the quinolone resistance-determining regions. Original mutated genes are indicated with an asterisk (Table 1). We identified 33, 36, 3, and 1 genetic variant of mexS, nfxB, gyrA, and gyrB, respectively. Genomic background and the presence of preexisting quinolone resistance mutations in gyrAB, nfxB, and mexS restrict early steps of ciprofloxacin resistance evolution (Table S3).
