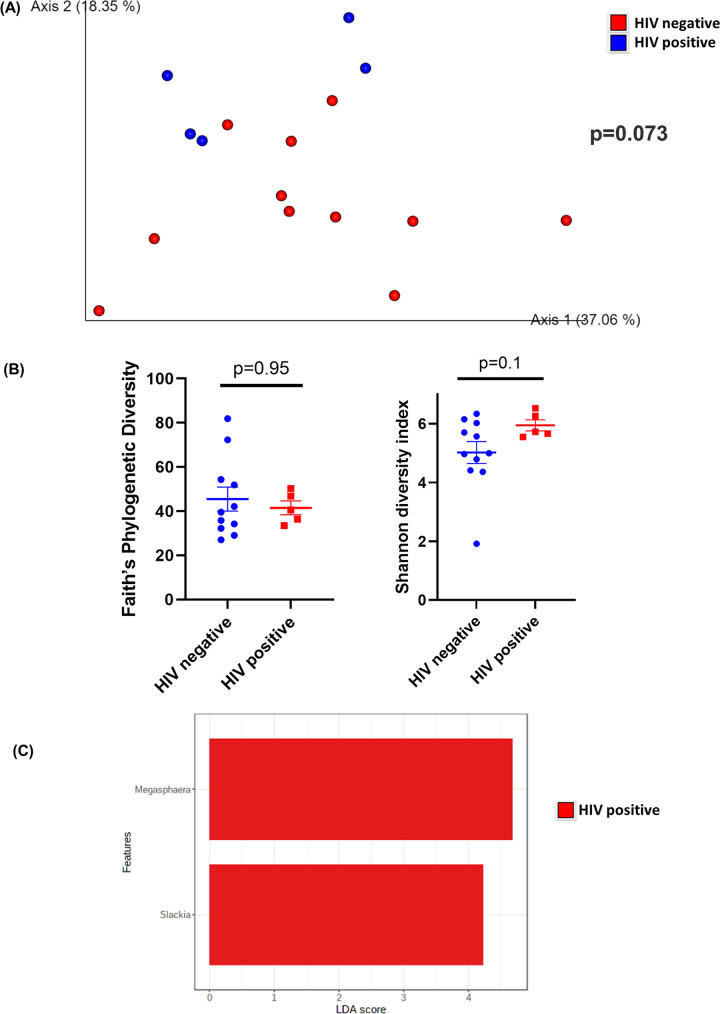
FIG 4.
Diversity and composition analysis of the colon brush samples. (A) Principal-coordinate analysis (PCoA) plot of weighted UniFrac distances (metrics of β-diversity). Samples grouped by HIV-negative (n = 12) and -positive (n = 5) status, P = 0.073. (B) Faith’s phylogenetic diversity and Shannon diversity index (metrics of α-diversity) at a sequencing depth of 80,000 reads. Samples were grouped by HIV-negative (n = 12) and -positive (n = 5) status. Error bars represent SEM. (C) Linear discriminant analysis effect size (LEfSe) analysis of top discriminative bacteria genera between gut samples from HIV-positive and -negative patients.