Figure 8.
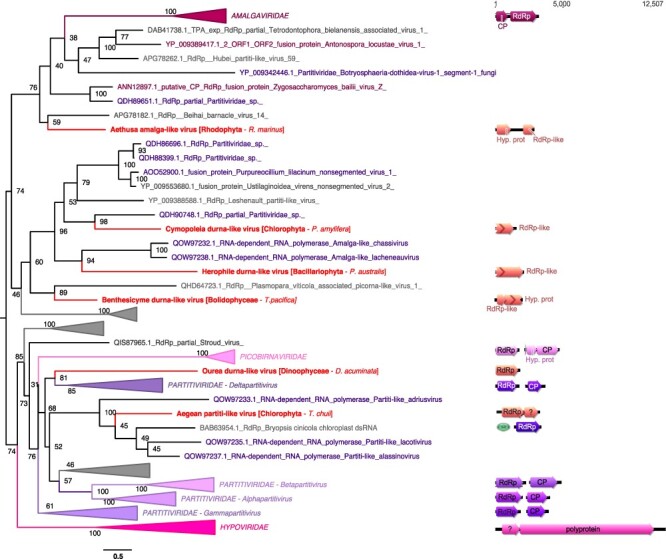
Phylogenetic positions of the newly described RNA viruses among the Durnavirales. Left, Maximum likelihood (ML) phylogeny of the Durnavirales RdRp (LG + F + R8 amino acid substitution model). Newly described viruses are indicated in red. Algae host taxon and species are specified in brackets. Branch labels = bootstrap support (%). The trees are mid-point rooted for clarity only. Right, genomic organization of newly discovered viruses (red), closest homologs, and the following Partiti-picobirna super-clade representatives: Zygosaccharomyces bailii virus Z (NC_003874; Amalgaviridae), Cryphonectria hypovirus 2 (NC_003534; Hypoviridae), Chicken picornavirus (NC_003534/NC_040438; Picobirnaviridae), Fig cryptic virus (NC_015494/NC_015495; Deltapartitivirus), Discula destructiva virus 1 (NC_002797/NC_002800; Gammapartitivirus), Ceratocystis resinifera virus 1 (NC_010755/NC_010754; Betapartitivirus), and White clover cryptic virus 1 (NC_006275/NC_006276; Alphapartitivirus). ORFs translated with the plastid genetic code are labelled with a green plastid. For clarity, some lineages were collapsed (a non-collapsed version of the tree is available as Supplementary Information).
