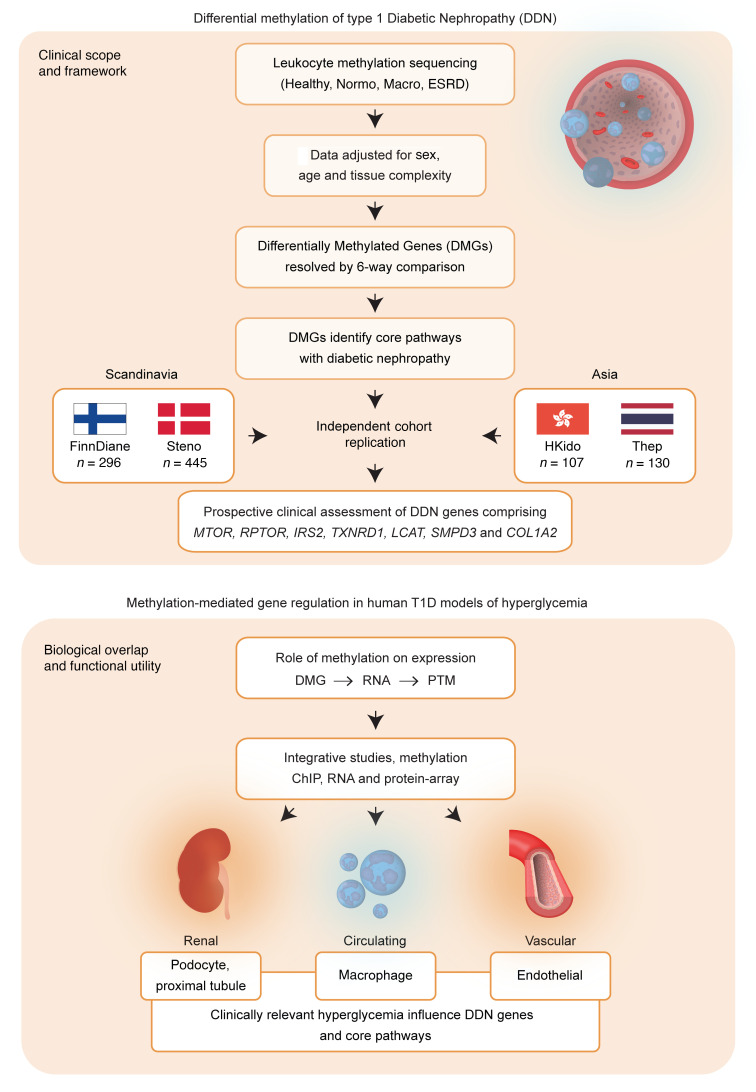
Figure 1. Clinical framework and experimental overlap.
The clinical scope and framework for determining differential methylation in type 1 diabetic nephropathy using Scandinavian and Asian cohorts. Genome-wide DNA methylation was analyzed to identify leukocyte-based DMRs in T1D participants from the Finnish Diabetic Nephropathy (FinnDiane) study using methyl-capture coupled with sequencing. DMRs were annotated to differentially methylated genes (DMGs) associated with progression of diabetic nephropathy (DDN) and validated in a larger subset of the FinnDiane study (n = 296) and independent cohorts from Denmark (n = 445), Hong Kong (107), and Thailand (n = 130). Biological overlap and functional utility were assessed for methylation-mediated gene regulation in human cell models of hyperglycemia. We performed functional studies in human podocytes, proximal convoluted tubule cells, macrophages, and vascular endothelial cells and confirmed methylation-mediated regulation of core genes MTOR, RPTOR, IRS2, COL1A2, TXNRD1, LCAT, and SMPD3. PTM, posttranslational modification.