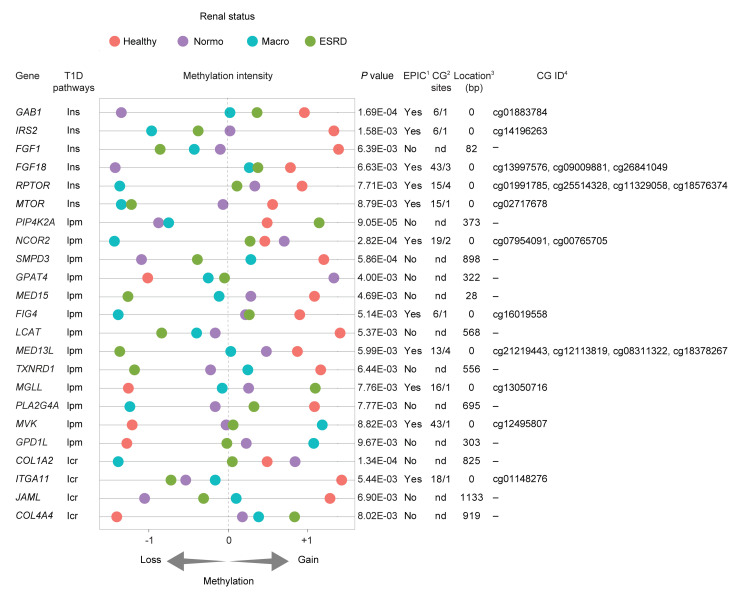
Figure 3. Functional analysis of FinnDiane cohort identified methylated genes associated with the progression of DN.
Differentially methylated genes associated with progression of diabetic nephropathy (DDN) with overlapping CTCF sites. Gene - annotated gene name; T1D-associated pathways with insulin signaling (Ins), lipid metabolism (lpm), and integrin-cell interaction (Icr); P value; Region length (bp); Methylation Intensity - scaled sequence abundance (–1, unmethylated; +1, methylated), showing relative gene methylation in healthy, Normo, Macro, and ESRD. Table also shows methyl-seq genes identified in the FinnDiane cohort intersected with probes from the Infinium Methylation EPIC profiling BeadChip microarray (EPIC array). Presence on the EPIC array; Number of CG sites assessed by methyl-seq overlapping EPIC array probe; Distance (bp) of the closest EPIC array probe from methyl-seq identified sequences. 1DMRs detected by methyl-seq–overlapping Infinium methylation EPIC BeadChip array probes. 2Comparison of CG number detected by methyl-seq versus EPIC probe CG location; nd denotes no detected probes on EPIC array. 3Distance of nearest EPIC array probe; 0 denotes EPIC CG site that exists within FinnDiane DMR. 4EPIC CG ID listed.