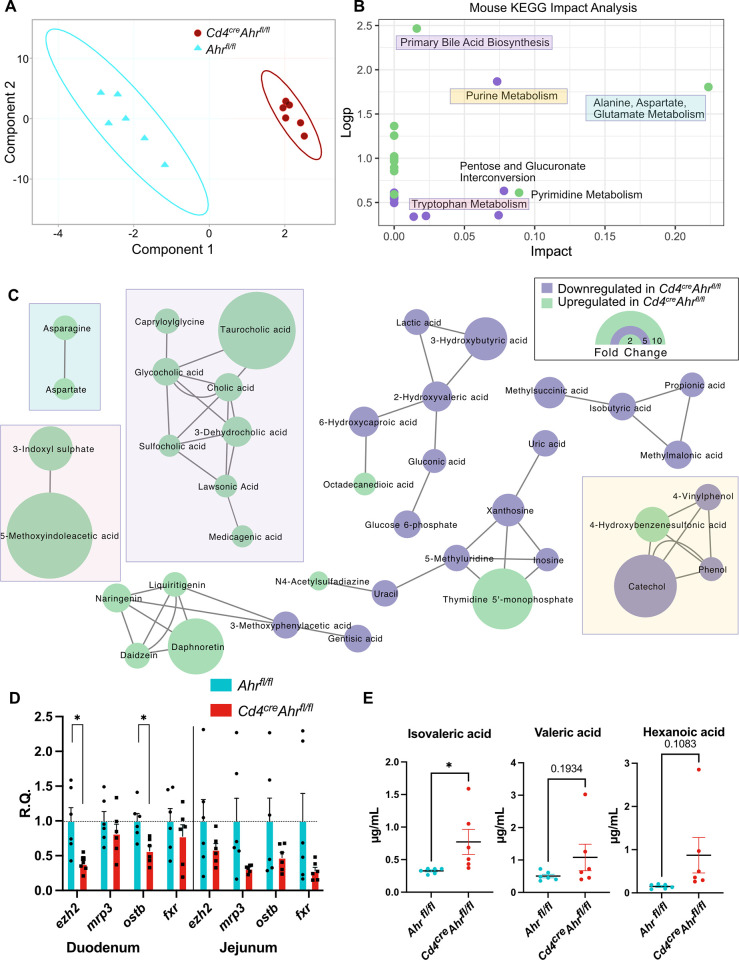
Fig 5. Elevated bile salts and SCFA detected in the cecum of AHR-deficient mice.
(A) Partial least-squares plot of LC-MS untargeted metabolomics of the <3 kDa fraction of the cecal contents from Cd4creAhrfl/fl and Ahrfl/fl mice. (B) Metaboanalyst mouse KEGG pathways significantly changed by genotype. (C) MetaMapp network analysis visualizing the chemically or biologically related significantly changed molecules. (D) qPCR of bile acid transporters (mrp3, ostb) and bile acid response elements (ezh2, fxr) in bulk tissue of the duodenum and jejunum of separately housed animals illustrated as relative quantity (R.Q.) from Ahrfl/fl tissue. (n = 6 mice/group; N = 2 experiments; multiple t tests with Welch correction [duodenum p = 0.0104, 0.3591, 0.0071, 0.3828; jejunum p = 0.2230, 0.0623, 0.1487, 0.1004]). (E) Quantitative targeted metabolomics for SCFAs show a significant increase in isovaleric acid and a trending increase in valeric and hexanoic acid in Cd4creAhrfl/f mice. (n = 6 mice/group; N = 1 experiment; unpaired t tests [p = 0.0427, 0.1934, 0.1083]) Error bars represent standard error from the mean. Raw data can be found in Supporting information (S1, S2, and S3 Data files). AHR, aryl hydrocarbon receptor; SCFA, short-chain fatty acid.