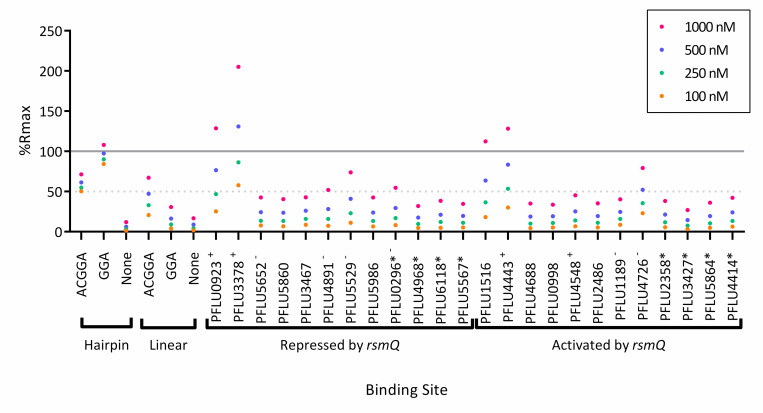
Fig 4. RsmQ binds to the upstream regions of predicted mRNA targets.
Purified RsmQ was tested against ssDNA oligos with synthetic oligos run as a control. Oligos are labelled as in S4 Table, with the genetic identifiers used to indicate the binding sites associated with those ORFs. Genes annotated with a * indicate ORFs that contain a simplified GGA binding site but are not predicted to have a full Rsm (AnGGA) binding site within 500 bp upstream or the first 100 bp of the ORF. Oligos predicted to have the full AnGGA (+) or a partial AnGGA (-) at the open top of a stem loop are indicated. Percentage Rmax values are shown for 4 concentrations. A solid line indicates 100% Rmax and a dotted line for 50% Rmax; 100% Rmax suggests 1:1 ligand protein binding as the experimentally acquired response is equal to the predicted response, with a 50% Rmax suggesting a weaker interaction or a 2:1/1:2 response. Each data point represents the mean of 2 replicates. Data are available in S3 Data. ssDNA, single-stranded DNA.