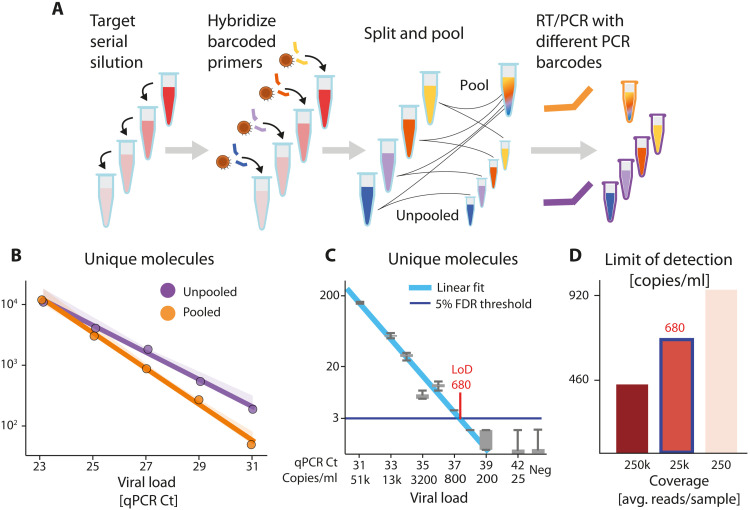
Fig. 4. ApharSeq sensitivity estimation on added samples.
(A) Target titration experimental scheme. SARS-CoV-2–positive sample is diluted in lysis buffer. Diluted samples are hybridized to barcoded ApharSeq primers (interior tube color) and split, so they can be assayed separately or pooled. Samples are subjected to PCR with different barcodes (purple/orange) to distinguish their treatments. (B) Assay is linear in pooled and unpooled ApharSeq assays (P < 0.001). The linearly extrapolated LoD for pooled samples is ~Ct 35.7 and is four times lower than the LoD for the single samples. (C) Low target titration experiment that includes addition of a viral target control for quantification. Gray boxes are molecular counts over read down-sampling replicates that show that the LoD is ~680 molecules/ml (Ct ~37.4). (D) LoD as a function of sequencing depth as derived by down-sampling the sequencing reads. Blue outlined bar (~25,000 reads per sample) corresponds to data in (C).