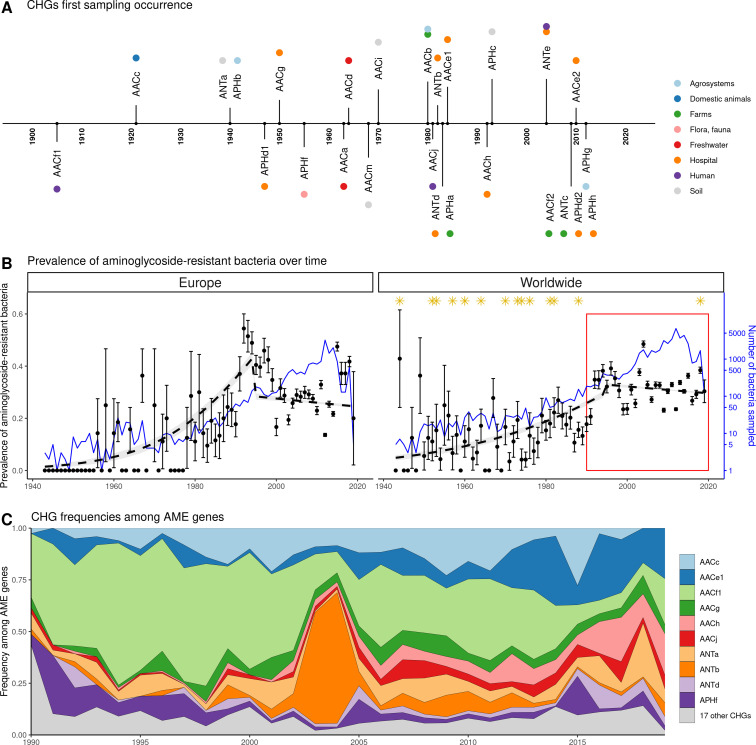
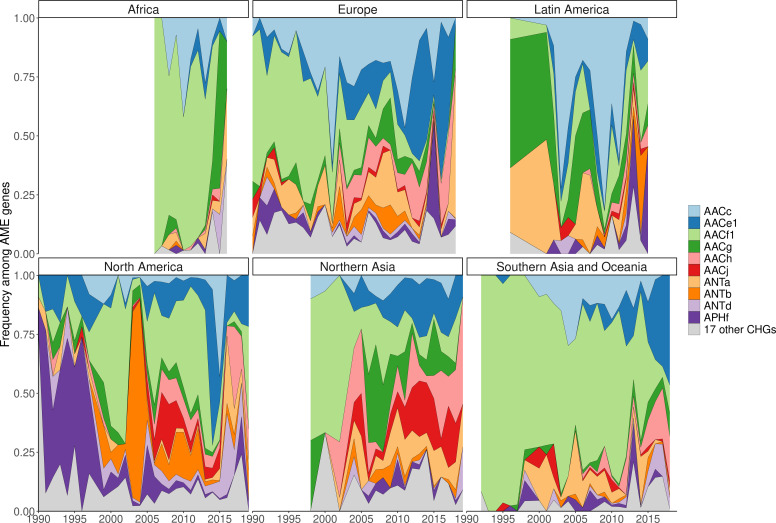
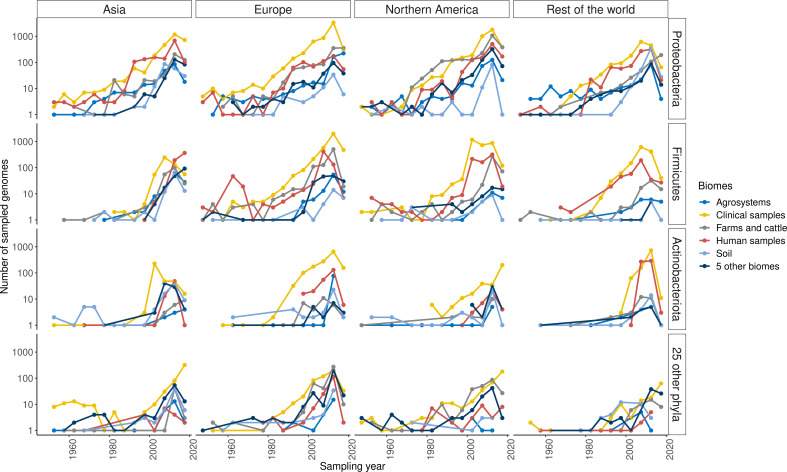
Figure 4. Global time trends for aminoglycoside resistance.
(A) First sampling occurrence for each CHG in the analyzed dataset. First sampling occurrence of APHe is unknown. (B) Evolution of the worldwide and European prevalences of AME bacteria between 1943 and 2019. The dots and error bars represent the measured prevalence (± standard error) of AME bacteria each year. The dotted line represents a binomial regression of the prevalence of AME bacteria over time, fitted separately before 1995 and after 1995. The blue curve represents the number of bacteria genomes sampled each year (in logarithmic scale). Orange stars represent the dates at which the following aminoglycoside drugs were released for commercial use: streptomycin, neomycin, hygromycin, kanamycin, paromomycin, capreomycin, gentamicin, ribostamycin, dibekacin, tobramycin, amikacin, sisomicin, netilmicin, micronomicin, isepamycin, plazomicin. The red rectangle displays the period of time analyzed in panel: (C) Evolution of worldwide CHG frequencies among sampled AME genes between 1990 and 2019. 17 CHGs for which less than 400 sequences were sampled are grouped and displayed in gray.