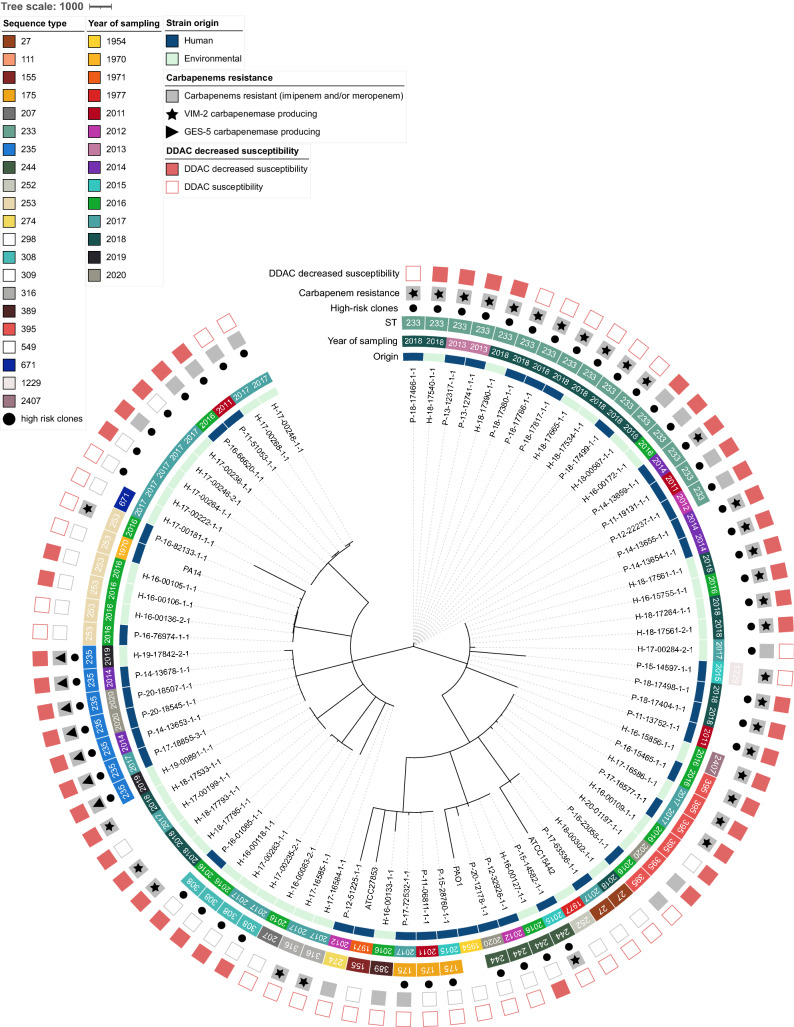
Figure 4.
Minimum spanning tree of 77 strains from panel for genomic characterization compared with reference strains (PAO1, PA14, ATCC15442 and ATCC27853). Core genome MLST clustering according to the cgMLST Pseudomonas aeruginosa scheme published by Tönnies et al.32. The tree was surrounded by the following information: the origin of the strains (patient, P or hospital environment, H), year of sampling, sequence type (ST), carbapenem resistance phenotype, whether it was associated with a VIM-2 or GES-5 carbapenemase, and decreased susceptibility phenotype to DDAC (didecyldimethylammonium chloride). GES-5: strains producing the class A carbapenemase GES-5; VIM-2: strains producing the class B carbapenemase VIM-2.