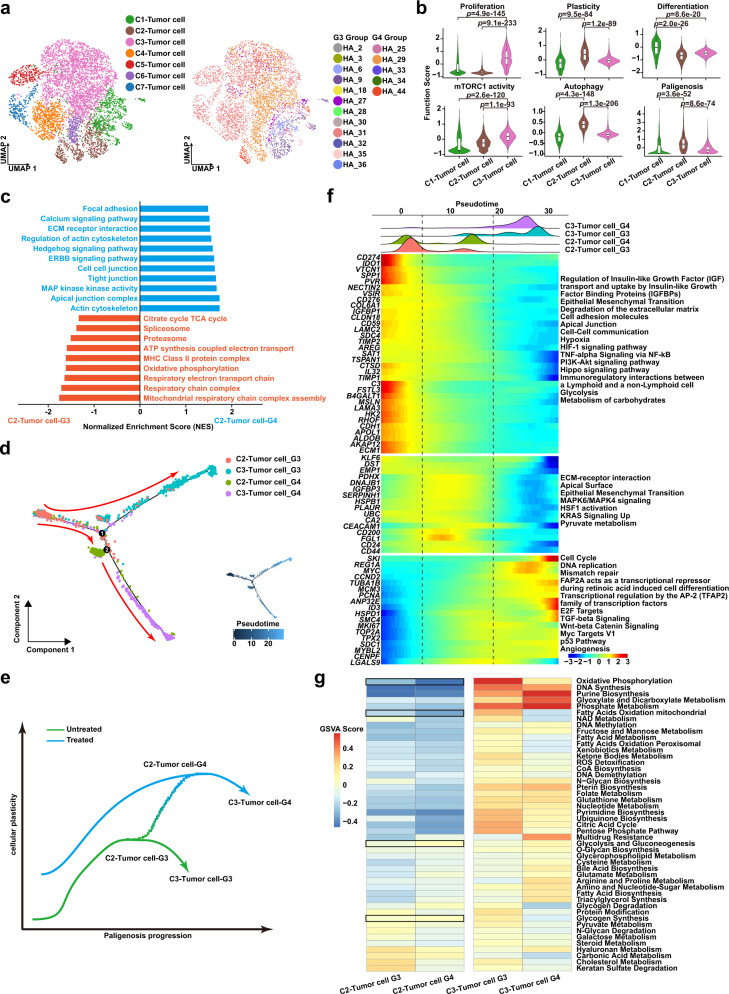
Fig. 6. High-plasticity GC evolves to high-proliferative GC through a conserved cellular program.
a Uniform Manifold Approximation and Projection (UMAP) visualization of tumor cells by cell types (Left, colors correspond to cell clusters) and samples (Right, colors correspond to samples). b Violin plot showing function scores of proliferation, plasticity, differentiation, mTORC1, autophagy, and paligenosis in tumor cells (C1-Tumor cell: n = 713 cells; C2-Tumor cell: n = 549 cells; C3-Tumor cell: n = 2545 cells). Box represents median ± interquartile range; whiskers represent 1.5x interquartile range; p-values are calculated by two-sided unpaired Wilcoxon test. c Gene Set Enrichment Analyses (GSEA) analysis showing distinct enrichment pathways of C2-Tumor cells in the G3 (red) and G4 Groups (blue). Bar chart showing the normalized enrichment score (NES) of specific pathways in specific tumor cells. d The trajectory plot of C2/C3-tumor cells in the G3 and G4 Groups (left), and the transition trajectories along pseudotime (right) in a two-dimensional state-space inferred by Monocle 2 analysis. e Two-dimensional schematic diagram showing cellular plasticity changes in C2/C3-tumor cells in the G3 and G4 Groups along paligenosis progression. f The 3-phase distribution of C2/C3-tumor cells along pseudotime color-coded by cell cluster (upper panel). Heatmap showing dynamic expression changes of genes and related pathways of C2/C3-tumor cells in the G3 and G4 Groups along pseudotime (lower panel). g Heatmap plot indicating the activity of metabolism pathways in C2/C3-tumor cells between the G3 and G4 Groups. Color represents the activity score of metabolism pathways calculated by Gene Set Variation Analysis (GSVA) analysis. Source data are provided as a Source Data file.