Figure 1.
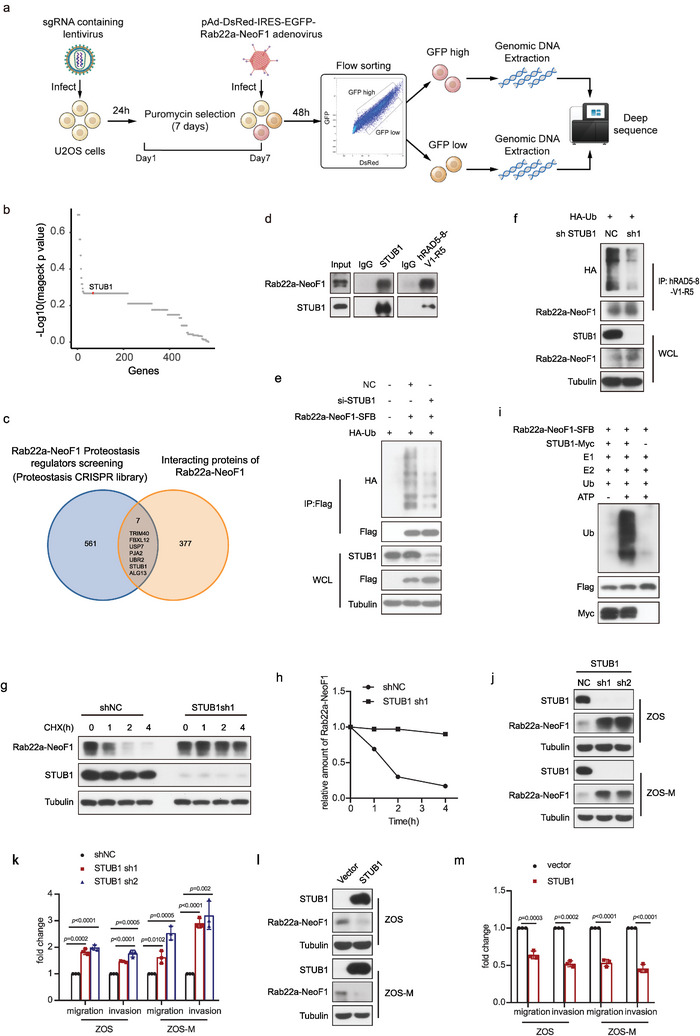
Flow cytometry‐based CRISPR‐Cas9 knockout library screening reveals STUB1 as an E3 ligase targeting Rab22a‐NeoF1 fusion protein in osteosarcoma. a) Schematic of CRISPR‐Cas9 knockout library screening pipeline and the gating strategy used in the screen. b) Scatterplot showing average log10 fold changes in mageck p value of sgRNA abundance in the GFP high‐sorted population. sgRNAs targeting STUB1 are highlighted in red. Each dot represents an average of log10 fold changes in mageck p values for ten independent sgRNAs per gene. c) Venn diagram indicates overlap of genes between the enrichment of sgRNAs targeting Rab22a‐NeoF1 in the CRISPR‐Cas9 knockout library screening and the interactome of Rab22a‐NeoF1 fusion protein by mass spectrometry. d) The co‐IPs were performed using ZOS‐M cells with IgG, hAb RAD5‐8, or anti‐STUB1 at their endogenous levels, and were analyzed by western blot. The experiments were repeated three times independently with similar results. e) Transfection of siRNAs targeting STUB1 or negative control (NC) in the 293T cells for 24 h and then the indicated plasmids were reinfected into these cells for 36 h. Cells were lysed and immunoprecipitated using anti‐FLAG beads followed by western blot analysis. The experiments were repeated three times independently with similar results. f) ZOS cells with knockdown of STUB1 by shRNAs or NC as indicated were transfected with HA‐Ubiquitin (HA‐Ub) for 48 h and then were lysed and immunoprecipitated using hRAD5‐8‐V1‐R5 antibody followed by western blot analysis. The experiments were repeated three times independently with similar results. g,h) ZOS cells with knockdown of STUB1 by shRNAs or shNC as indicated were treated with g) cycloheximide (CHX: 20 µg mL−1) for the indicated time and subjected to western blotting analysis. h) Quantitation of the Rab22a‐NeoF1 protein levels in (g). The experiments were repeated three times independently with similar results. i) The indicated Rab22a‐NeoF1‐SFB and STUB1‐Myc protein were affinity‐isolated using streptavidin‐conjugated beads and anti‐Myc affinity gel, respectively, from lysates of HEK293T cells transfected with SFB‐tagged Rab22a‐NeoF1 or MYC‐tagged STUB1 for 48 h, and then were subjected to the in vitro ubiquitination assay. The experiments were repeated three times independently with similar results. j,l) The indicated stable ZOS or ZOS‐M cells were analyzed by western blot. The experiments were repeated three times independently with similar results. k,m) Quantification analyses of migration and invasion assays using the indicated stable ZOS or ZOS‐M cells. The depicted results are the averages of at least three independent experiments. The data are presented as the mean ± SD. A two‐sided unpaired student's t‐test was performed, and p values are shown.
