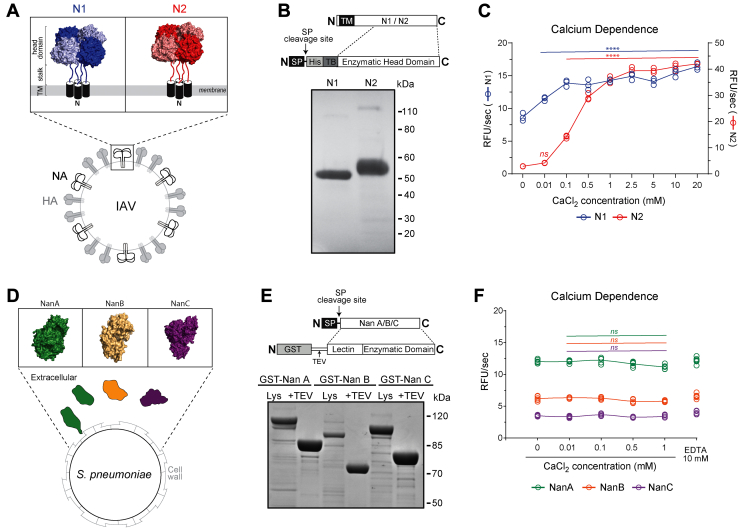
Figure 1.
Recombinant neuraminidase isolation and calcium dependence.A, diagram showing the topology and structure of neuraminidase subtypes 1 (N1) and 2 (N2) from seasonal (H1N1 and H3N2) IAVs. Structures of the tetrameric N1 (PDB ID: 3NSS) (76) and N2 (PDB ID: 4K1K) (77) head domains were generated with PyMol. B, recombinant N1 and N2 (∼5 μg each) were resolved under reducing conditions by SDS-PAGE (4–12% gel) and visualized by Coomassie staining. The N1 and N2 head domain constructs inserted into the baculovirus genome for insect cell expression are displayed above the gel. Regions corresponding to the transmembrane domain (TM), signal peptide (SP), 6× His tag (His), and the tetramerization domain (TB) are indicated. C, activity of the recombinant N1 and N2 in the presence of the indicated CaCl2 concentrations was determined using MUNANA. p values were calculated by a one-way ANOVA Dunnett’s multiple comparisons test with a 95% CI using 0 mM calcium as the comparison condition to characterize an increase in activity due to calcium addition. ∗∗∗∗p < 0.0001. ns – not significant. D, diagram showing the topology and structure of the three neuraminidases (NanA, NanB, and NanC) encoded by pneumococci. For NanA both the cell wall–associated and secreted variant are shown. PyMol was used to generate the NanA (PDB ID: 2YA4) (78), NanB (PDB ID: 2VW0) (43) and NanC (PDB ID: 4YZ1) (79) monomer structures. E, recombinant NanA, NanB, and NanC (∼5 μg each) were resolved under reducing conditions by SDS-PAGE (4–12% gel) and visualized by Coomassie staining. The constructs expressed in E. coli are displayed above the gel. Regions corresponding to glutathione S-transferase (GST), TEV protease site (TEV), and the signal peptide (SP) are indicated. F, recombinant NanA, NanB, and NanC activities in the presence of CaCl2 and EDTA were determined using MUNANA. IAV, influenza A viruses; MUNANA, 4-methylumbelliferyl-α-D-N-acetylneuraminide.