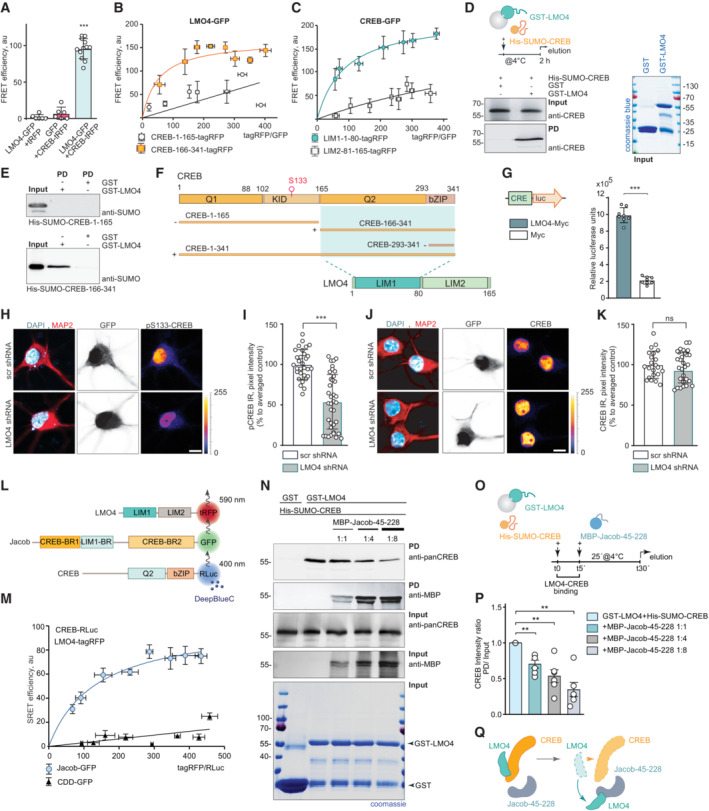
Figure 3. Jacob can displace LMO4 from cAMP‐responsive element‐binding protein (CREB).
-
AFluorescence resonance energy transfer (FRET) measurements indicate a tight association between LMO4‐GFP and CREB‐tagRFP. N = 6–12 independent experiments measured in triplicates.
-
B, CFRET saturation experiments with (B) LMO4‐GFP and CREB‐1‐165‐tagRFP or (C) CREB‐166‐341‐tagRFP and CREB‐GFP and LIM1‐1‐80‐tagRFP or LIM2‐81–165‐tagRFP revealed an association between the LIM1 domain of LMO4 and the C‐terminus of CREB. N = 8 independent experiments.
-
DRecombinant His‐SUMO‐CREB directly binds to GST‐LMO4 in pull‐down experiments.
-
EThe interaction between CREB with LMO4 is mediated by the C‐terminus of CREB (166‐341 aa), but not by its N‐terminus (1‐165 aa), as evidenced by pull‐down experiments between recombinant His‐SUMO‐1‐165‐CREB, His‐SUMO‐166‐341‐CREB, and GST‐LMO4.
-
FSchematic representation of CREB and LMO4 domain structure and fusion constructs used for the experiments. Light green boxes represent the interaction interface.
-
GMyc‐LMO4 overexpression increases CREB‐dependent luciferase expression in HEK293T cells expressing luciferase under the CRE promoter. Relative luciferase units in cells overexpressing Myc‐LMO4 as compared to Myc‐transfected controls. N = 8 from two independent experiments.
-
H–KKnockdown of LMO4 reduces nuclear pCREB immunoreactivity. (H, J) Representative confocal images of hippocampal neurons transfected with LMO4 shRNA construct or scrambled control (both expressing GFP under CMV promoter as a transfection control). Scale bar: 10 μm. Dot plots representing the mean of nuclear (I) pCREB or (K) CREB staining intensity normalized to scrambled control. N = 30–37 nuclei analyzed from at least 3 independent cell cultures.
-
L, M(L) Schematic representation of constructs used in SRET experiments. “BR” stands for binding region. (M) SRET saturation experiments reveal that Jacob‐GFP forms a triple complex with CREB‐RLuc and LMO4‐tagRFP. A caldendrin (CDD‐GFP) construct was used as negative control. N = 8 independent experiments.
-
N–QThe N‐terminus of Jacob displaces LMO4 from CREB. (N) GST‐LMO4 coupled to beads was preincubated with His‐SUMO‐CREB and subsequently incubated with an increasing amount of MBP‐Jacob‐45‐228. (O) Schematic depicts the timeline of the competition pull‐down experiment. (P) Bar graphs represent six independent experiments per condition.
Data information: (H, J) Lookup table indicates the pixel intensities from 0 to 255. ns, non‐significant difference, **P < 0.01, ***P < 0.001 by (G, I, K) two‐tailed Student's t‐test or (P) one‐sample t‐test or (A) one‐way ANOVA followed by Bonferroni's multiple comparisons test. All data are represented as mean ± SEM.
Source data are available online for this figure.
