Figure EV3. Nitarsone disrupts the Jacob‐LMO4 interaction.
-
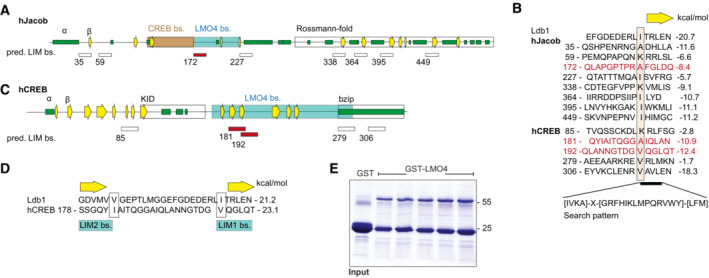
A–DPredicted binding sites for LMO4 LIM domains in Jacob and cAMP‐responsive element‐binding protein (CREB). (A) Schematic structure of human Jacob showing predicted secondary structures (helices, green; β‐strands, yellow arrows) and experimentally determined binding regions for CREB (orange) and LMO4 (gray). The C‐terminus of Jacob is predicted to have a Rossmann‐fold similar to caspases. (B) The LIM1 binding peptide of Ldb1 is aligned to 8 sequences of Jacob that match the search pattern for the conserved hydrophobic residues and the adjacent β‐strand. Structures of LIM1:peptides were modeled and free energy ΔΔG were calculated. Only the peptide starting at 172 (red) lies within the LMO4 binding region. In human, CREB 5 matching peptides were identified. (C) Schematic structure of human CREB with labeled LMO4‐binding region and known KID and bZIP domains. (D) The two peptides starting at 181 and 192 are within the LMO4‐binding region and align to Ldb1 peptide where 181 binds to LIM2 and 192 to LIM1.
-
EImage of gels stained with coomassie blue showing the purity of bacterially produced GST‐LMO4 used for pull‐down assay.
