Abstract
Seven new compounds, including three isocoumarins (1–3), three pyrrolidinone derivatives (8–10), and one pentaene diacid (15), together with 13 known compounds, were isolated from the rice culture of the endophytic fungus Fusarium decemcellulare F25. Their structures and stereochemistry were established using HRESIMS, NMR, electronic circular dichroism (ECD) calculations, and single-crystal X-ray diffraction. The possible biosynthetic pathways for compounds 1–3 and 8–10 were proposed. The antifungal efficacies of compounds 1 – 20 were evaluated against Colletotrichum musae, and compounds 13, 14, and 17 exhibited inhibitory activities against C. musae with MIC values of 256, 64 and 128 μg/mL, respectively.
Keywords: Fusarium decemcellulare F25, secondary metabolites, isocoumarins, pyrrolidinones, antifungal activities
1. Introduction
Endophytic fungi are microorganisms that asymptomatically colonize living tissues of healthy plants (Li and Lou, 2018). Their complex interactions with the plant host, other organisms, and the external environment result in the production of secondary metabolites that are often characterized by diverse structures and biological activities (Zhang et al., 2006; Gao et al., 2018; Gupta et al., 2019).
Mahonia fortunei is a traditional Chinese medicinal plant, and its root, stem, and leave can be used as medicine for treating bacterial infection, pneumoconiosis, psoriasis, and cough (Zhang et al., 2015). Fungal endophytes from this medicinal plant have afforded many bioactive novel natural products, indicating that mining fungi from this host is an effective strategy for obtaining potential lead compounds (Li et al., 2015, 2016; Wang et al., 2019; Tian et al., 2021). Specifically, an antibacterial tetracyclic triterpenoid with a unique aromatic B-ring, and a cytochalasan with a new 6/6/5-fused tricyclic core skeleton were isolated from M. fortunei-derived endophytic fungi (Li et al., 2015; Wang et al., 2019).
In our continuous research on fungal endophytes from M. fortunei, endophytic Fusarium decemcellulare F25 was obtained. Study on secondary metabolites from F. decemcellulare is relatively less. Li et al. reported three cyclic pentapeptides and an antifungal cyclic lipopeptide from an endophytic fungus, F. decemcellulare LG53 (Li et al., 2016). The well-known shikimic acid can be produced by F. decemcellulare harboring in the fruits of the plant Flacourtia inermis (Qader et al., 2018). Under guidance of 1H NMR, 12 polypropionate derivatives were isolated from a marine-derived fungus F. decemcellulare SYSUMS6716, and two compounds, decempyrones C and J, exhibited potent anti-inflammatory activity and inhibitory activity against protein tyrosine phosphatase A (Guo et al., 2021).
In the specific ecological niche, endophytic fungi could coevolve with associated organisms, such as other endophytic fungi and environmental pathogens. This usually made fungal endophytes produce antifungal compounds for chemical defense (Li et al., 2015). Therefore, the antifungal activity of the ethyl acetate (EtOAc) extract of F. decemcellulare F25 was investigated. Its ethyl acetate extract showed a significant inhibition activity against Colletotrichum musae ACCC 31244, revealing the production of bioactive metabolites and being worth for chemical investigation.
Further isolation led to the identification of three new isocoumarins (1–3), three new pyrrolidinones (8–10), and one new pentaene diacid (15), together with 13 known compounds from the endophytic fungus F. decemcellulare F25 (Figure 1). Herein, we report their isolation, identification, and biological activity, together with the proposed biosynthetic pathway.
Figure 1.
Structures of compounds 1 – 20.
2. Materials and methods
2.1. General experimental procedures
Optical rotations were acquired on a JASCOP-1020 polarimeter. ECD data were measured on a Chirascan spectropolarimeter. IR spectra were measured on PerkinElmer infrared spectrophotometer. 1D and 2D NMR spectra were recorded on Bruker Avance 400 or 600 DRX spectrometers in acetone-d6, methanol-d4, DMSO-d6 and chloroform-d. Column chromatography (CC) was performed on silica gel (200–300 mesh; Qingdao Marine Chemical Plant Branch., China), RP-C18 (ODS-A, 50 μm, YMC, Kyoto, Japan), or Sephadex LH-20 (100–200 mesh; Beijing Solarbio Technology Co., Ltd., China). Plates precoated with silica gel GF254 (Rushan, Shandong Sun Desiccant Co., Ltd.) were used for thin layer chromatography (TLC). An Agilent HPLC series 1260 and Shimadzu LC-20AR were used for analysis and isolation. For analysis, an Agilent Eclipse XDB-C18 column (4.6 × 150 mm, 5 μm) was used. The isolation was achieved on an Agilent semi-preparative Eclipse XDB-C18 column (9.4 × 250 mm, 5 μm). HPLC-MS data were acquired on an Agilent 1260 Series system coupled with an Agilent Accurate-Mass-Q-TOF MS 6520 system equipped with an Electrospray ionization (ESI) source.
2.2. Fungal material
The endophytic fungus F. decemcellulare F25 was isolated from the stem of the Chinese medicinal plant M. fortunei collected from Qingdao, People’s Republic of China. The fungal strain was deposited in 20% glycerol at −80°C in the school of Pharmaceutical Sciences, Guizhou University, Guizhou, China. The endophytic fungus was identified as F. decemcellulare by the analysis of internal transcribed spacer (ITS) region of the rDNA (GenBank No. OQ001346).
2.3. Fermentation and extraction
The fungal strain F. decemcellulare F25 was cultured on potato dextrose agar (PDA) media for a week at 28 ± 2°C. A week-old culture plate was cut into small pieces under aseptic conditions, and were then inoculated into 394 flasks (300 mL) each containing 40 g of rice, 0.12 g of peptone, and 60 mL of water. The cultures were incubated at 28 ± 2°C for 40 days. Afterward, the whole cultures were extracted with ethyl acetate by sonication under ice bath conditions for three times. Then the EtOAc solution was collected and evaporated to dryness, affording 352.2 g of brown extracts. After suspension of the crude extract in water, petroleum ether and EtOAc were used to extract 294.6 g and 50.0 g of the corresponding organic phase, respectively.
2.4. Isolation and purification
The EtOAc extract was fractionated by column chromatography (CC) on ODS eluting with a gradient of acetonitrile (CH3CN)/H2O (0:100; 3:7; 5:5; 7:3; 100:0, v/v, each 8 L) to give eight fractions (Fr. A–Fr. H).
Fraction A was applied to semi-preparative HPLC to yield compounds 11 (tR 22.1 min, 53.3 mg), 4 (tR 38.2 min, 8.9 mg), 5 (tR 39.1 min, 16.5 mg), 6 (tR 40.3 min, 29.1 mg), and a mixture of 8 and 9 (tR 29.2 min, 16.2 mg), eluting with a gradient of CH3CN in H2O from 40 to 65%. The above mixture was subjected to isolation on a chiral HPLC column to afford compounds 8 (tR 25.1 min, 6.2 mg) and 9 (tR 28.9 min, 5.9 mg). Fraction A (14.4 g) was also separated into 12 subfractions (A1 − A12) by CC on silica gel eluted by CH2Cl2/CH3OH (1:0, 40:1, 30:1, 20:1, 10:1, 5:1 and 0:1, v/v, each 7 L). Subfraction A3 was purified by semi-preparative HPLC with a gradient elution from 30 to 85% CH3CN in H2O to afford compounds 1 (tR 33.0 min, 11.5 mg) and 18 (tR 12.6 min, 16.4 mg). Subfraction A6 was then applied to semi-preparative HPLC with a gradient from 30 to 55% CH3CN in H2O as eluent to obtain compound 19 (tR 14.2 min, 25.3 mg). Compound 10 (tR 33 min, 27.8 mg) was purified by semi-preparative HPLC, eluting with a gradient of CH3CN in H2O from 40 to 65% (v/v) as eluent from subfraction A7. Compound 2 (tR 17.5 min, 10.1 mg) was obtained from subfraction A8 using semi-preparative HPLC with a gradient elution from 30 to 85% CH3CN in H2O. Subfrcation A9 was separated by semi-preparative HPLC with a gradient elution from 30 to 75% CH3CN in H2O to yield compound 3 (tR 27.0 min, 10.6 mg).
Fraction C was fractionated by CC on Sephadex LH-20 eluting with CH3OH/CH2Cl2 (1:1, v/v) to give five subfractions (C1 – C5). Subfraction C3 was purified by semi-preparative HPLC with a gradient elution from 45 to 50% CH3CN in H2O with 0.1% trifluoroacetic acid (TFA) to afford compounds 15 (tR 38.1 min, 7.5 mg) and 16 (tR 37.5 min, 10.3 mg). Fraction C4 was subjected to semi-preparative HPLC with a gradient elution from 35 to 65% CH3CN in H2O to yield compound 7 (tR 16.0 min, 2.0 mg).
Fraction D was purified by semi-preparative HPLC eluting with gradient from 60 to 70% CH3CN in H2O with 0.1% TFA to offer compound 17 (tR 33.5 min, 6.3 mg). Compound 12 (tR 27.2 min, 6.2 mg) was obtained from fraction F by semi-preparative HPLC eluting with a gradient elution from 50 to 100% CH3CN in H2O. Compounds 13 (tR 19.3 min, 17.5 mg), 14 (tR 22.0 min, 17.7 mg), and 20 (tR 28.5 min, 13.1 mg) were obtained from fraction G using semi-preparative HPLC with a gradient elution from 75 to 100% CH3CN in H2O.
2.5. Spectroscopic data of compounds
Compound (1), yellowish solid; LC-UV (CH3CN in H2O) λmax: 248, 330 nm; IR νmax: 3319, 2942, 2832, 1677, 1020 cm−1; 1H NMR (DMSO-d6, 400 MHz); and 13C NMR (DMSO-d6, 100 MHz) data, see Table 1; HRESIMS m/z 285.0762 [M + H]+ (calcd. For C16H13O5, 285.0757).
Table 1.
1H NMR (400 MHz, δ in ppm) and 13C NMR Data (100 MHz, δ in ppm) of 1 – 3 (DMSO-d6).
| Position | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δC, type | δH (J in Hz) | δC, type | δH (J in Hz) | δC, type | δH (J in Hz) | |
| 1 | 166.0, C | 165.5, C | 166,2, C | |||
| 3 | 150.8, C | 154.9, C | 151.7, C | |||
| 4 | 108.7, CH | 6.69, s | 105.9, CH | 6.55, s | 108.9, CH | 6.67, s |
| 4a | 138.2, C | 139.3, C | 139.6, C | |||
| 5 | 103.3, CH | 6.47,d (2.1) | 103.3, CH | 6.57, d (2.3) | 103.8, CH | 6.72, d (2.2) |
| 6 | 165.7, C | 164.2, C | 164.3, C | |||
| 7 | 101.9, CH | 6.38, d (2.1) | 102.5, CH | 6.64, d (2.3) | 102.8, CH | 6.61, d (2.2) |
| 8 | 162.7, C | 162.2, C | 162.3, C | |||
| 8a | 98.4, C | 99.9, C | 100.1, C | |||
| 9 | 120.0, C | 40.9, CH2 | 2.60, m | 111.5, C | ||
| 10 | 155.9, C | 64.6, CH | 4.28, m | 157.3, C | ||
| 11 | 113.0, CH | 6.75, d (8.3) | 50.3, CH2 | 2.57, m | 100.3, CH | 6.23, d (2.2) |
| 12 | 130.6, CH | 7.18, t (8.3) | 207.7, C | 159.3,C | ||
| 13 | 120.6, CH | 6.77, d (8.3) | 30.5, CH3 | 2.11, s | 108.4,CH | 6.19, d (2.2) |
| 14 | 139.6, C | 139.3, C | ||||
| 15 | 19.5, CH3 | 2.21, s | 19.9, CH3 | 2.12, s | ||
| 1’ | 100.0, CH | 5.74, d (4.4) | 100.0, CH | 5.76, d (4.5) | ||
| 2’ | 71.5, CH | 4.10, m | 71.6, CH | 4.11, m | ||
| 3’ | 69.2, CH | 3.96, m | 69.3, CH | 3.93, m | ||
| 4’ | 86.7, CH | 3.96, m | 86.9, CH | 3.98, m | ||
| 5’ | 61.4, CH2 | 3.48, m | 61.5, CH2 | 3.49, m | ||
Compound (2), yellowish solid; +51.6 (c 0.68, CH3OH); LC-UV (CH3CN in H2O) λmax: 244, 276, 330 nm; IR νmax: 3330, 2925, 1681, 1642, 1625, 1572, 1357, 1237, 1163, 1018, 990 cm−1; 1H NMR (DMSO-d6, 400 MHz); and 13C NMR (DMSO-d6,100 MHz) data, see Table 1; HRESIMS m/z 411.1289 [M + H]+ (calcd. For C19H23O10, 411.1286).
Compound (3), yellowish solid; +63.2 (c 0.56, CH3OH); LC-UV (CH3CN in H2O) λmax: 204, 236, 336 nm; IR νmax: 3310, 2917, 1678, 1616, 1570, 1505, 1470, 1400, 1158, 1024, 999 cm−1; 1H NMR (DMSO-d6, 400 MHz); and 13C NMR (DMSO-d6,100 MHz) data, see Table 1; HRESIMS m/z 433.1133 [M + H]+ (calcd. For C21H21O10, 433.1129).
Compound (8), colorless oil; -22.7 (c 0.59, CH3OH); LC-UV (CH3CN in H2O) λmax: 200, 230, 286 nm; IR νmax: 3329, 2946, 2836, 1661, 1451, 1408, 1114, 1017 cm−1 1H NMR (DMSO-d6, 400 MHz) and 13C NMR (DMSO-d6,100 MHz) data, see Supplementary Table 1; HRESIMS m/z 334,1,649 [M + H]+ (calcd. For C18H24NO5, 334.1649).
Compound (9), colorless oil; +8.23 (c 0.61, CH3OH); LC-UV (CH3CN in H2O) λmax: 200, 230, 286 nm; IR νmax: 3329, 2946, 2836, 1661, 1451, 1408, 1114, 1017 cm−1;1H NMR (DMSO-d6, 400 MHz); and 13C NMR (DMSO-d6,100 MHz) data, see Supplementary Table 1; HRESIMS m/z 334, 1,652 [M + H]+ (calcd. For C18H24NO5, 334.1649).
Compound (10), colorless oil; -24.1 (c 2.25, CH3OH); LC-UV (CH3CN in H2O) λmax: 204, 228, 286 nm; IR νmax: 3336, 3286, 2905, 1644, 1427, 1367, 1334, 1314, 1164, 1053, 1030 cm−1; 1H NMR (DMSO-d6, 400 MHz); and 13C NMR (DMSO-d6,100 MHz) data, see Supplementary Table 1; HRESIMS m/z 336,1808 [M + H]+ (calcd. For C18H26NO5, 336.1805).
Compound (15), yellow powder; LC-UV (CH3CN in H2O) λmax: 220, 280, 360 nm; IR νmax: 3387, 2833, 1698, 1475, 1391, 1017 cm−1; 1H NMR (DMSO-d6, 400 MHz); and 13C NMR (DMSO-d6,100 MHz) data, see Supplementary Table 2; HRESIMS m/z 263.1277 [M + H]+ (calcd. For C15H19O4, 263.1278).
2.6. X-ray crystallographic analysis of compound 9
The crystal structure of compound 9 was obtained from the solution of CH3OH. A suitable crystal were collected on a Bruker APEX-II CCD Venture diffractometer using graphite-monochromated Mo Kα radiation (λ = 0.710 73 Å) at 297 K. Absorption correction using equivalent reflctions was performed with the SADABS program. Crystallographic tables were constructed using Olex2 (Dolomanov et al., 2010). The structure was solved with the Shelxt software package
(Sheldrick, 2015), and refined with the Shelxt refinement package using Least Squares minimization.
Crystal data for compound 9: C18H25NO6 (M = 351.39 g/mol): triclinic, space group P1, a = 6.4597(7) Å, b = 7.5258(7) Å, c = 9.4129(7) Å, α = 95°, β = 100°, γ = 93°, V = 93.981(8) Å3, Z = 1, T = 297 K, μ(Mo Kα) = 0.098 mm−1, F(000) = 188, ρcalc = 1.311 g/cm3; of the 8,205 reflections measured (4.44° ≤ 2Θ ≤ 50.01°), 2,823 were unique (Rint = 0.0922, Rsigma = 0.0810) which used in all calculations. The final R1 was 0.0596 (I > 2σ(I)), and wR2 was 0.1706 (all data).
2.7. Antifungal assay
Following our previously established methods (Wang et al., 2019; Tian et al., 2021), the crude extract of F. decemcellulare F25 was firstly evaluated for antifungal activity against five plant pathogens (Colletotrichum musae ACCC 31244, Alternaria solani, Fusarium foetens, Fusarium mangiferae, and Lasiodiplodia pseudotheobromae) by agar diffusion assay. The crude extract showed inhibitory activity against C. musae ACCC 31244, indicating the production of antifungal molecules. Further antifungal evaluation of pure compounds against C. musae ACCC 31244 was determined with the broth dilution method, and provided minimum inhibitory concentration (MIC) values. The cycloheximide was used as a positive control in parallel to reveal the comparative antifungal efficacy of compounds 1–20.
3. Results and discussion
3.1. OSMAC screen and fermentation of Fusarium decemcellulare F25
The OSMAC (One Strain Many Compounds) approach refers to the activation of many silent gene clusters in microorganisms by altering the culture environment of the strain. This strategy maximizes the biosynthetic capacity of a microorganism that produces structurally diverse and biologically active secondary metabolites. The F. decemcellulare F25 was cultured on four different solid media including rice-based, soybean-based, corn-based, and czapek-dox agar (CDA) culture. Remarkably, HPLC chromatograms showed a number of peaks in rice-based culture, suggesting that the rice medium strongly triggered the production of secondary metabolites (Supplementary Figure 1).
3.2. Screening of antifungal activities of crude extract
The antifungal activities of crude extracts were evaluated against five plant pathogens. Compared with the positive control drug cycloheximide, it was found that, at the concentration of 40 μg/paper disk, the crude extract of F. decemcellulare F25 showed antifungal activity against the fungal C. musae (Supplementary Figure 2). Considering the abundant secondary metabolites and the antifungal activity of F. decemcellulare F25, this strain F25 was further subjected to chemical investigation.
3.3. Structural characterization of these isolated compounds
Seven new compounds, including three isocoumarins (1–3), three pyrrolidinone derivatives (8–10), and one pentaene diacid (15), together with 13 known compounds, were isolated from the rice culture of the endophytic fungus Fusarium decemcellulare F25.
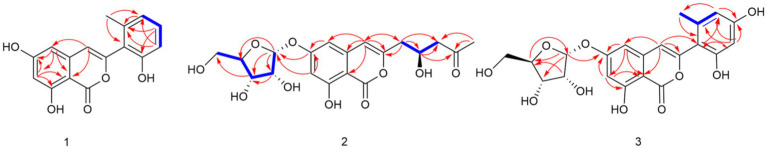
Compound 1 was obtained as a yellowish solid. Its molecular formula C16H12O5 was established by the HRESIMS at m/z 285.0762 [M + H]+ (calcd. For 285.0757), implying eleven degrees of unsaturation. The presence of hydroxyl and carbonyl groups were implied by IR absorption bands at 3319 and 1677 cm−1, respectively. The 1H and 13C NMR data of 1 (Table 1) were highly similar to those of pleosporalone A (Cao et al., 2016), excepted for a proton at C-7 in 1 rather than a methyl group in pleosporalone A. In addition, the coupling constant between H-5 and H-7 (J = 2.1 Hz) proved that there is no methyl substitution at C-7 of 1. The HMBC correlations (Figure 2) from H-5 (δH 6.47) to C-6 (δC 165.7), C-7 (δC 101.9), and C-8a (δC 98.4), and from H-7 (δH 6.38) to C-5 (δC 103.3), C-6 (δC 165.7), C-8 (δC 162.7), and C-8a (δC 98.4), together with the HSQC spectrum (δC/δH 101.9/6.38 and 103.3/6.47 ppm) further confirmed that there is a proton attached to C-7. Final detailed analysis of the HSQC and HMBC spectra allowed the assignment for all proton and carbon resonances of 1. Thus, the structure of compound 1 was assigned completely.
Figure 2.
Key COSY (bold lines) and HMBC (arrows) correlations of compounds 1–3.
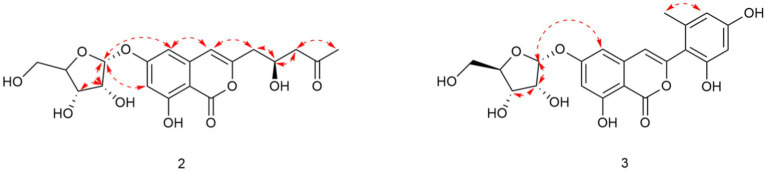
Compound 2 was obtained as a yellowish solid. Its molecular formula C19H22O10 was established by the HRESIMS at m/z 411.1289 [M + H]+ (calcd for 411.1286), implying nine degrees of unsaturation. The presence of hydroxyl and carbonyl groups were shown by IR absorption bands at 3330, 1681 cm−1, respectively. The attachment of sugar unit was determined to be ribose by comparison of 1H and 13C NMR data of compound 2 with those of daldiniside C. The coupling constant of the anomeric proton at δH 5.74 (1H, d, J = 4.4 Hz) indicated the ribose unit should be α-configured (Hu et al., 2014). Additionally, the pentose moiety was linked to C-6 proved by the correlation of H-1’/C-6 observed in the HMBC (Figure 2) experiment and H-1’/H-5, H-1’/H-7 in NOESY spectrum (Figure 3). The 1H NMR and 13C NMR data (Table 1) showed the presence of an isocoumarin unit in 2, whose structure was the same as that of compound 6 ((−)-citreoisocoumarin)) (Yamamura et al., 1991) by comparison with spectroscopic data. Thus, the structure of compound 2 was established.
Figure 3.
Key NOESY correlations of compounds 2 and 3.
Compound 3 was obtained as a yellowish solid. Its molecular formula C21H20O10 was established by the HRESIMS at m/z 433.1133 [M + H]+ (calcd for 433.1129), implying 12 degrees of unsaturation. The presence of hydroxyl and carbonyl groups were shown by IR absorption bands at 3310, 1678 cm−1, respectively. The 1H and 13C NMR data of aglycone in 3 (Table 1) were highly similar to those of polyisocoumarin (Jiang et al., 2020), which was previously isolated from Polygonum cuspidatum. However, an α-ribose attached at the C-6 of 3 rather than a β-d-glucopyrancose at C-6 in polyisocoumarin. And the NMR data showed that compound 3 and compound 2 have the same ribose at C-6. Thus, the structure of compound 3 was assigned completely.
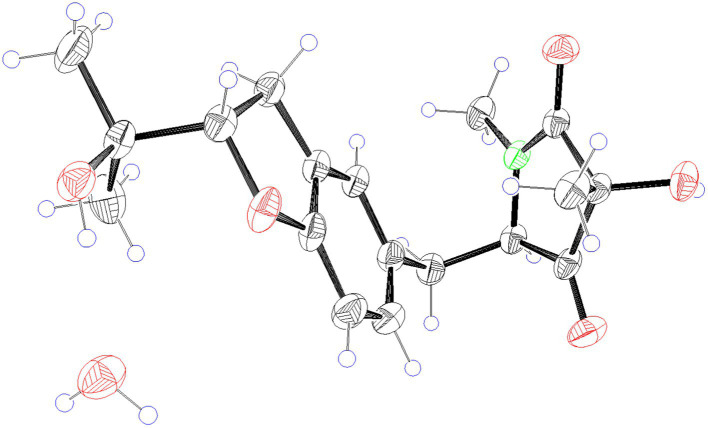
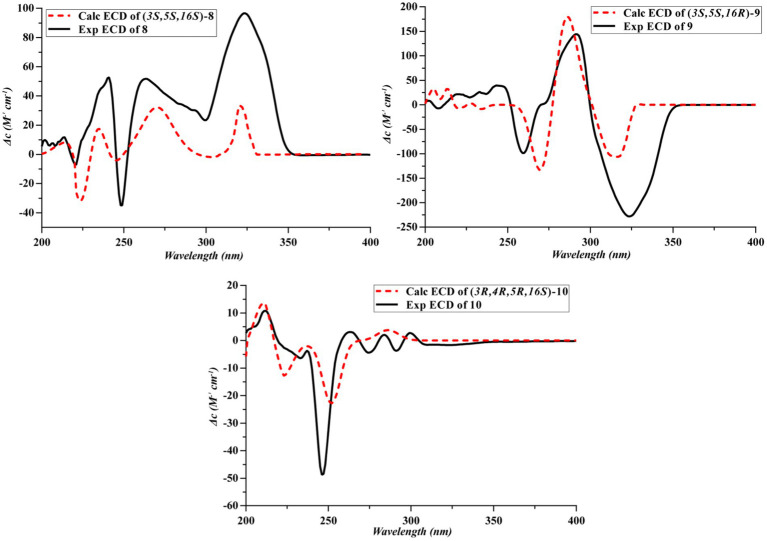
Compounds 8 and 9 were obtained as colorless oil. Their molecular formula C18H23NO5 were established by the HRESIMS at m/z 334.1649 and 334.1652 [M + H] +, respectively (calcd for 334.1649), implying eight degrees of unsaturation. The presence of hydroxyl and carbonyl groups were implied by IR absorption bands at 3329 and 1661 cm−1, respectively. Detailed analysis of the 1H and 13C NMR data suggested that 8 possessed the same planar structure as that of rigidiusculamide C (Li et al., 2009). A comparison of the NMR data (Supplementary Table 1) of 8 and 9 suggested that they differed only in the substituent at C-16. Detailed analysis of the HSQC and HMBC spectra allowed the assignment for all proton and carbon resonances of 9. The relative configuration of 9 was assigned by single-crystal X-ray diffraction as shown in Figure 4. To clarify the absolute configurations of 8 and 9, ECD calculations were performed by the time dependent density functional theory-predicted curve calculated at the quantum mechanical level. The calculated electronic circular dichroism (ECD) curve of (3S, 5S, 16S)-8 matched well with the 8 experimental ECD data, and the ECD curve of (3S, 5S, 16R)-9 is in good agreement with the experimental ECD data of 9. Therefore, the absolute configuration of compound 8 was determined as 3S, 5S, 16S, and the absolute configuration of compound 9 was 3S, 5S, 16R (Figure 5).
Figure 4.
X-ray ORTEP drawing of compound 9.
Figure 5.
Experimental and calculated ECD spectra of 8–10.
Compound 10 was obtained as a yellow oil. Its molecular formula C18H25NO5 was established by the HRESIMS at m/z 336.1808 [M + H]+ (calcd for 336.1805), implying seven degrees of unsaturation. The presence of hydroxyl and carbonyl groups were implied by IR absorption bands at 3336 and 1644 cm−1, respectively. The NMR data (Supplementary Table 1) of 10 revealed highly similar identical structural features to those found in 8, except that the C-4 ketone carbon was replaced by an oxygenated methine in 10. And 10 has the same planar structure as rigidiusculamide D (Li et al., 2009). The NOESY correlation of CH3 with H-4 indicates that 3-OH and 4-OH are located on the same face, whereas H-5 and H-4 placed on the opposite face of the ring supported by the absence of a NOESY correlation between H-5 and H-4. To determine the absolute configuration, the simulated electronic circular dichroism (ECD) spectra of 10 were obtained from the calculation of Gaussian 16 based on time-dependent density functional theory. The absolute configurations of C-3, C-4, C-5, and C-16 in 10 were deduced as 3R, 4R, 5R, and 16S, respectively, by comparing calculated ECD spectra with the experimental ECD spectrum (Figure 5).
Compound 15 was isolated as a yellow powder. Its molecular formula C15H18O4 was established by the HRESIMS at m/z 263.1277 [M + H]+ (calcd for 263.1278), implying seven degrees of unsaturation. The presence of hydroxyl and carbonyl groups were implied by IR absorption bands at 3387 and 1698 cm−1, respectively. A comparison of NMR data (Supplementary Table 2) with those of nectriacid C (Cui et al., 2016) suggested that 15 possessed a closely similar structure as nectriacid C, except that there is no methoxy group (δH 3.74, δC 51.7) in 15. Its NMR data suggested that 15 belonged to the pentaene diacid derivative. Thus, the structure of compound 15 was assigned completely.
The remaining 13 known compounds from the F. decemcellulare F25 were identified as 12-epicitreoisocoumarinol (4) (Cui et al., 2016), eoisocoumarinol (5) (Cui et al., 2016), (−)-citreoisocoumarin (6) (Yamamura et al., 1991; Ola et al., 2013), trichophenol A (7) (Liu et al., 2020), rigidiusculamide B (11) (Li et al., 2009), fusaristatins A (12) (Shiono et al., 2007), enniatin H (13) (Nilanonta et al., 2003), enniatin I (14) (Nilanonta et al., 2003), nectriacid A (16) (Cui et al., 2016), nectriacid B (17) (Cui et al., 2016), 4-hydroxy-3,6-dimethyl-2 H-pyrane-2-one (18) (Hirota et al., 1999), macrocarpon C (19), (Ola et al., 2013), and α-linoleic acid (20) (Zeng et al., 2017) by comparison of their MS and NMR data with those reported in the literature.
3.4. Antifungal assays
Compounds 1–20 were assayed for their antifungal activities. The results showed that compounds 13, 14, and 17 exhibited inhibitory activities against the plant-pathogenic fungus C. musae ACCC31244 with MICs of 256, 64, and 128 μg/mL, respectively. The MIC of the positive control cycliheximide was 32 μg/mL.
3.5. Plausible biogenetic pathways
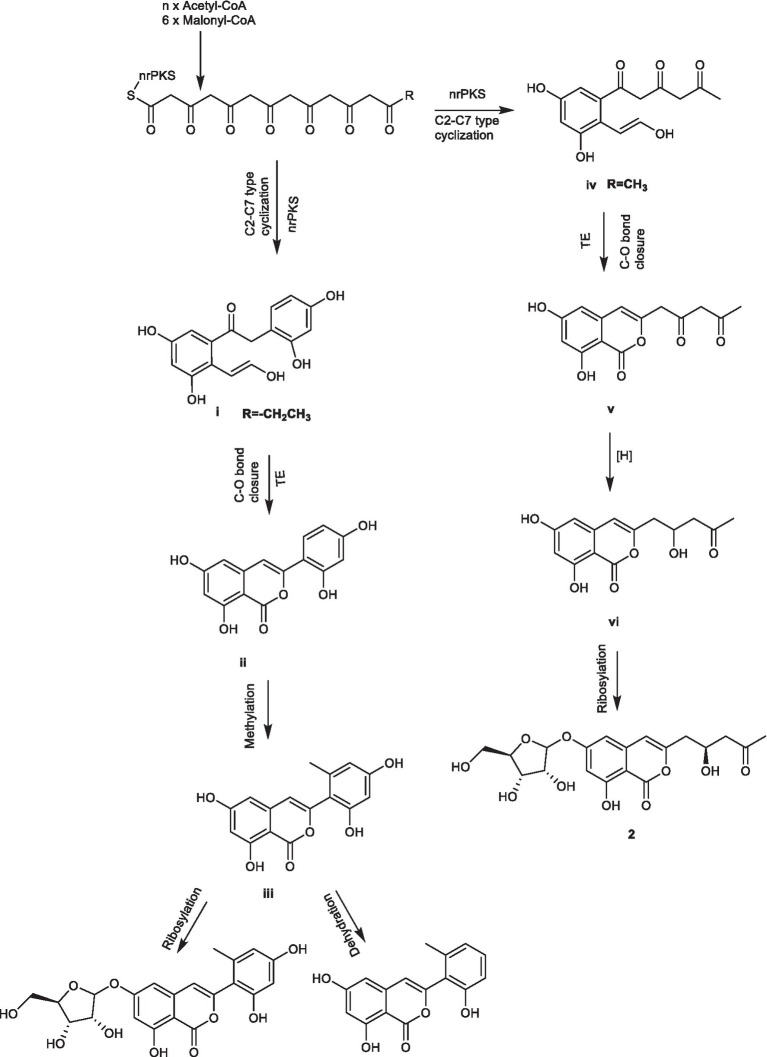
The biosynthetic pathways of compounds 1 and 3 (Scheme 1) start with condensation catalyzed by nonreducing polyketide synthase (nrPKS) of two acetyl-coenzyme A molecules and six malonyl-CoA molecules resulting in the formation of the intermediate i (Liu and Begley, 2020). Intermediate i was catalyzed by TE domains or spontaneous C-O bond closure to form intermediate ii. The isocoumarin 1 was derived from intermediate ii through methylation and dehydration, while, intermediate ii will create 3 by methylation and ribosylation. The biosynthetic pathway of 2 (Scheme 1) starts with condensation catalyzed by a modular (nrPKS) of one acetyl-coenzyme A molecule and six malonyl-CoA molecules resulting in the formation of the intermediate iv. Intermediate iv was catalyzed by thioesterase (TE) domains or spontaneous C − O bond closure to form intermediate v. A series of reductive modifications for this intermediate led to intermediate vi. Intermediate vi then underwent a ribosylation reaction to afford 2.
SCHEME 1.
Plausible biogenetic pathways of compounds 1 – 3.
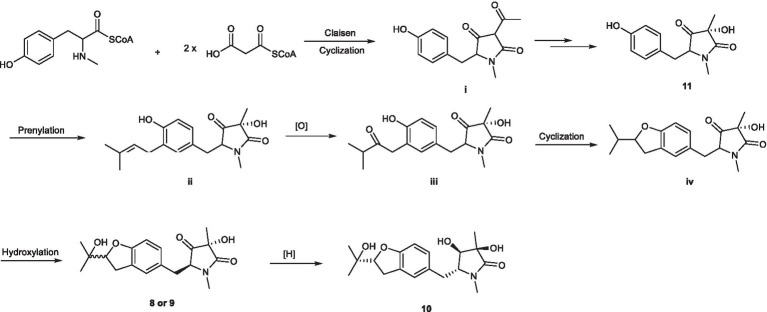
As shown in Scheme 2, pyrrolidones originate from the cyclization of an amino acid and a polyketide (Royles, 1995; Li et al., 2009), leading to the formation of 11. The pyrrolidones 8 and 9 were likely to be biogenetically derived from 11 through prenylation, oxidation, cyclization, and hydroxylation. The derivative 10 was formed from 8 or 9 through one-step hydrogenation.
SCHEME 2.
Plausible biogenetic pathways of compounds 8 – 10.
4. Conclusion
In summary, the chemical investigation on the endophytic fungus F. decemcellulare F25 resulted in the isolation and identification of twenty secondary metabolites, including three new isocoumarin derivatives (1–3), three new pyrrolidinones (8–10), and one new pentaene diacid (15), together with thirteen known compounds. Compounds 13, 14, and 17 exhibited antifungal activities against plant pathogen C. musae ACCC31244. This study reveals the potential of endophytic fungi as a promising source of bioactive compounds.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: GenBank No. OQ001346 of ITS region of the rDNA in NCBI and Deposition Number 2232225 of X-ray crystallographic data in CCDC.
Author contributions
ZS and KZ: conception or design. KZ, ZS, YJS, GL, SX, and CY: acquisition, analysis, or interpretation of data. ZS, KZ, GL, and CY: drafting the work or revising and final approval of the manuscript. All authors have reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
We gratefully acknowledge the National Natural Science Foundation of China (grant nos. 22067002 and 82060635), the Science and Technology Foundation of Guizhou (grant no. J[2020]1Y049), and Guizhou University (grant no. (2018)04) for the financial supports.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1127971/full#supplementary-material
References
- Cao F., Yang J. K., Liu Y. F., Zhu H. J., Wang C. Y. (2016). Pleosporalone a, the first azaphilone characterized with aromatic A-ring from a marine-derived Pleosporales sp. fungus. Nat. Prod. Res. 30, 2448–2452. doi: 10.1080/14786419.2016.1198352, PMID: [DOI] [PubMed] [Google Scholar]
- Cui H., Liu Y., Nie Y., Liu Z., Chen S., Zhang Z., et al. (2016). Polyketides from the mangrove-derived endophytic fungus Nectria sp. HN001 and their α-glucosidase inhibitory activity. Mar. Drugs 14, 1660–3397. doi: 10.3390/md14050086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolomanov O. V., Bourhis L. J., Gildea R. J., Howard J. A. K., Puschmann H. (2010). OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Cryst. 42, 339–341. doi: 10.1107/S0021889808042726 [DOI] [Google Scholar]
- Gao H., Li G., Lou H. X. (2018). Structural diversity and biological activities of novel secondary metabolites from endophytes. Molecules 23, 646–677. doi: 10.3390/molecules23030646, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Wu Q., Chen D., Jiang M., Chen B., et al. (2021). Absolute configuration of polypropionate derivatives: Decempyrones A–J and their MptpA inhibition and anti-inflammatory activities. Bioorg. Chem. 115:105156. doi: 10.1016/j.bioorg.2021.105156, PMID: [DOI] [PubMed] [Google Scholar]
- Gupta S., Chaturvedi P., Kulkarni M. G., Staden J. V. (2019). A critical review on exploiting the pharmaceutical potential of plant endophytic fungi. Biotechnol. Adv. 39:107462. doi: 10.1016/j.bioteschadv.2019.107462, PMID: [DOI] [PubMed] [Google Scholar]
- Hirota A., Nemoto A., Tsuchiya Y., Hojo H., Abe N. (1999). Isolation of a 2-pyrone compound as an antioxidant from a fungus and its new reaction product with 1, 1-diphenyl-2-picrylhydrazyl radical. Biosci. Biotechnol. Biochem. 63, 418–420. doi: 10.1271/bbb.63.418, PMID: [DOI] [PubMed] [Google Scholar]
- Hu Z. X., Xue Y. B., Bi X. B., Zhang J. W., Luo Z. W., Li X. N., et al. (2014). Five new secondary metabolites produced by a marine-associated fungus, Daldinia eschscholzii. Mar. Drugs 12, 5563–5575. doi: 10.3390/md12115563, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. S., Li F. S., Feng Z. M., Yang Y. N., Zhang P. C. (2020). New phenolic glycosides from Polygonum cuspidatum. J. Asian Nat. Prod. Res. 22, 17–23. doi: 10.1080/10286020.2019.1646730, PMID: [DOI] [PubMed] [Google Scholar]
- Li G., Kusari S., Golz C., Strohmann C., Spiteller M. (2016). Three cyclic pentapeptides and a cyclic lipopeptide produced by endophytic Fusarium decemcellulare LG53. RSC Adv. 6, 54092–54098. doi: 10.1039/C6RA10905E [DOI] [Google Scholar]
- Li G., Kusari S., Kusari P., Kayser O., Spiteller M. (2015). Endophytic Diaporthe sp. LG23 produces a potent antibacterial tetracyclic triterpenoid. J. Nat. Prod. 78, 2128–2132. doi: 10.1021/acs.jnatprod.5b00170, PMID: [DOI] [PubMed] [Google Scholar]
- Li J., Liu S., Niu S., Zhuang W., Che Y. (2009). Pyrrolidinones from the ascomycete fungus Albonectria rigidiuscula. J. Nat. Prod. 72, 2184–2187. doi: 10.1021/np900619z, PMID: [DOI] [PubMed] [Google Scholar]
- Li G., Lou H. X. (2018). Strategies to diversify natural products for drug discovery. Med. Res. Rev. 38, 1255–1294. doi: 10.1002/med.21474, PMID: [DOI] [PubMed] [Google Scholar]
- Liu H. W., Begley T. P. (2020). Comprehensive Natural Products III. Oxford: Elsevier. [Google Scholar]
- Liu X. H., Hou X. L., Song Y. P., Wang B. G., Ji N. Y. (2020). Cyclonerane sesquiterpenes and an isocoumarin derivative from the marine-alga-endophytic fungus Trichoderma citrinoviride A-WH-20-3. Fitoterapia 141:104469. doi: 10.1016/j.fitote.2020.104469, PMID: [DOI] [PubMed] [Google Scholar]
- Nilanonta C., Isaka M., Chanphen R., Thong-Orn N., Tanticharoen M., Thebtaranonth Y. (2003). Unusual enniatins produced by the insect pathogenic fungus Verticillium hemipterigenum: isolation and studies on precursor-directed biosynthesis. Tetrahedron 59, 1015–1020. doi: 10.1016/S0040-4020(02)01631-9 [DOI] [Google Scholar]
- Ola A. R. B., Thomy D., Lai D., Brötz-Oesterhelt H., Proksch P. (2013). Inducing secondary metabolite production by the endophytic fungus Fusarium tricinctum through coculture with Bacillus subtilis. J. Nat. Prod. 76, 2094–2099. doi: 10.1021/np400589h, PMID: [DOI] [PubMed] [Google Scholar]
- Qader M. M., Kumar N. S., Jayasinghe L., Fujimoto Y. (2018). Shikimic acid production by Fusarium decemcellulare, an endophytic fungus isolated from Flacourtia inermis fruits. J. Biol. Active Prod. Nat. 8, 43–50. doi: 10.1080/22311866.2018.1426494 [DOI] [Google Scholar]
- Royles B. J. L. (1995). Naturally occurring tetramic acids: structure, isolation, and synthesis. Chem. Rev. 95, 1981–2001. doi: 10.1021/cr00038a009 [DOI] [Google Scholar]
- Sheldrick G. M. (2015). SHELXT–integrated space-group and crystal-structure determination. Acta Cryst 71, 3–8. doi: 10.1107/S2053273314026370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiono Y., Tsuchinari M., Shimanuki K., Miyajima T., Murayama T., Koseki T., et al. (2007). Fusaristatins a and B, two new cyclic lipopeptides from an endophytic Fusarium sp. J. Antibiot. 60, 309–316. doi: 10.1038/ja.2007.39, PMID: [DOI] [PubMed] [Google Scholar]
- Tian C., Gao H., Peng X. P., Li G., Lou H. X. (2021). Fusidic acid derivatives from the endophytic fungus Acremonium pilosum F47. J. Asian Nat. Prod. Res. 23, 1148–1155. doi: 10.1080/10286020.2020.1866559, PMID: [DOI] [PubMed] [Google Scholar]
- Wang H. H., Li G., Qiao Y. N., Sun Y., Peng X. P., Lou H. X. (2019). Chamiside a, a cytochalasan with a tricyclic core skeleton from the endophytic fungus Chaetomium nigricolor F5. Org. Lett. 21, 3319–3322. doi: 10.1021/acs.orglett.9b01065, PMID: [DOI] [PubMed] [Google Scholar]
- Yamamura S., Lai S., Shizuri Y., Kawai K., Furukawa H. (1991). Three new phenolic metalolites from Penicillium species. Heterocycles 32, 297–305. doi: 10.3987/COM-90-5639 [DOI] [Google Scholar]
- Zeng J. X., Bing X. B., Ying B. I., Wang J., Ren G., Wang H. L., et al. (2017). Chemical constitutes from plantaginis semen(II). Chin. J. Exp. Tradit. Med. Form. 23, 81–84. doi: 10.13422/j.cnki.syfjx.2017040081 [DOI] [Google Scholar]
- Zhang H. W., Song Y. C., Tan R. X. (2006). Biology and chemistry of endophytes. Nat. Prod. Rep. 23, 753–711. doi: 10.1039/b609472b [DOI] [PubMed] [Google Scholar]
- Zhang D., Tao X., Chen R., Liu J., Li L., Fang X., et al. (2015). Pericoannosin a, a polyketide synthase-nonribosomal peptide synthetase hybrid metabolite with new carbon skeleton from the endophytic fungus Periconia sp. Org. Lett. 17, 4304–4307. doi: 10.1021/acs.orglett.5b02123, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: GenBank No. OQ001346 of ITS region of the rDNA in NCBI and Deposition Number 2232225 of X-ray crystallographic data in CCDC.