Abstract
Exosomes are the phospholipid-membrane-bound subpopulation of extracellular vesicles derived from the plasma membrane. The main activity of exosomes is cellular communication. In cancer, exosomes play an important rolefrom two distinct perspectives, one related to carcinogenesis and the other as theragnostic and drug delivery tools. The outer phospholipid membrane of Exosome improves drug targeting efficiency. . Some of the vital features of exosomes such as biocompatibility, low toxicity, and low immunogenicity make it a more exciting drug delivery system. Exosome-based drug delivery is a new innovative approach to cancer treatment. Exosome-associated biomarker analysis heralded a new era of cancer diagnostics in a more specific way. This Review focuses on exosome biogenesis, sources, isolation, interrelationship with cancer and exosome-related cancer biomarkers, drug loading methods, exosome-based biomolecule delivery, advances and limitations of exosome-based drug delivery, and exosome-based drug delivery in clinical settings studies. The exosome-based understanding of cancer will change the diagnostic and therapeutic approach in the future.
Keywords: Eexosomes, cancer, drug loading methods, drug delivery, cancer biomarker
1. Introduction
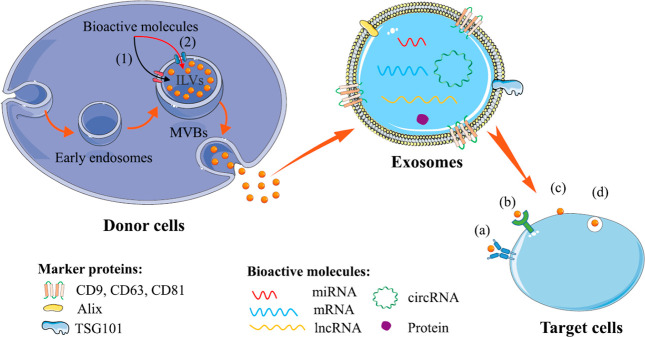
Exosomes are nanoscale extracellular vesicles secreted from several cells.1,2 This is the most fast-growing research field. The most interesting thing about the exosome is that it is the messenger of several pathological conditions. The fundamental level is involved in cellular communication.3 It transports several biologically active cargoes, for example, DNA,4 RNA,2,5 proteins,6−8 etc. This cargo can transform the cellular behavior of uptaking recipient cells. Cancer and exosomes have the most thrilling association. The collective evidence shows that tumor-derived exosomes (TEXs) regulate cell signaling and reprogramming in the complex tumor microenvironment (TME) to promote cancer development (uncontrolled cell growth, angiogenesis, metastasis, organ-specific metastasis immune evasion, and drug resistance).9−11 TEXs carry the molecular signature to help the early detection of cancer and work as biomarkers of cancer. Multiple nanodrug delivery technologies are being studied to improve medication potency, minimize toxicity, increase efficacy, and prolong drug flux duration. Early endosomes first develop when endocytic vesicles on the plasma membrane protrude outward. After changing into late endosomes, the early endosomes start to build up intraluminal vesicles (ILVs) in their lumen. This happens when the endocytic membrane enlarges inward. Endosomes ILVs are frequently referred to as MVBs due to their outward appearance. One group of bioactive molecules integrated into ILVs during MVB synthesis includes proteins, mRNA, miRNA, lncRNA, and circRNA (Figure 112).
Figure 1.
Overview of exosome biogenesis via (1) the ESCRT-dependent pathway and (2) the ESCRT-independent pathway involving exosome biogenesis and cargo selection of molecules. Target cells uptake exosomes via different pathways such as (a) antigen presentation, (b) cell signaling, (c) cell membrane fusion, and (d) pinocytosis or phagocytosis. Reproduced with permission from ref (12). Copyright 2020 Elsevier.
Exosome formation and biological cargo selection and loading are regulated via (1) the ESCRT-dependent process and (2) the ESCRT-independent pathway. ILVs are eventually discharged as exosomes into the extracellular environment when MVBs fuse with the plasma membrane. Several mechanisms, including (a) antigen presentation, (b) cell signaling, (c) cell membrane fusion, and (d) pinocytosis or phagocytosis, might lead to the uptake of these exosomes by target cells. In the drug delivery sector, natural or synthetic polymers and liposomes are more explored members. Both efficient drug delivery systems have several limitations, for example, low stability, toxicity, and low biocompatibility.13 In this crisis, exosomes show a promising role in drug delivery in in vivo and in vitro systems.14 Exosomes are overcoming all limitations of polymers and liposomes, which is the reason why they are becoming the brightest star in the drug delivery research area.13,15 In this review, we will cover exosome biogenesis, exosome sources, the exosome isolation process, the interrelation between exosomes and cancer, exosome drug loading methods, and the application of exosomes against several cancers and finally highlight the clinical study related to exosome-based drug delivery.
2. Biogenesis of Exosomes
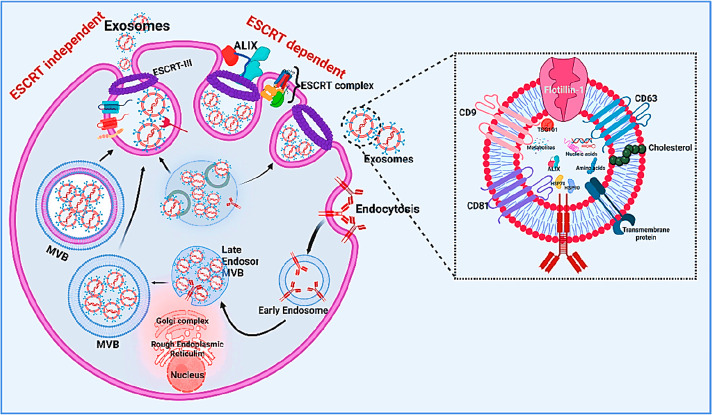
Exosomes are dynamic entities continuously generated from the endosomal system within the cell and exposed to the extracellular environment through the process of exocytosis. The membrane of the multivesicular body (MVB) invaginates to form the late endosomal system, further elongating the late endosomes in the fold to form intraluminal vesicles (ILVs).16 During the formation of the ILVs, some specific proteins are incorporated into the vesicles, and these vesicles fuse with the perimeter or plasma membrane of the cell; these vesicles are termed exosomes.17 An interesting point about the structures of exosomes is that they are cup-shaped or biconcave when artificially produced by drying but in solution appear spherical when observed under the transmission electron microscope.18 There are many reports from the previous literature that some of the intricate protein machinery contributes to the formation of ILVs. This protein complex is termed the transport-required endosomal sorting complex, or ESCRT.19 Four different ESCRT subunits (0, I, II, and III) play key roles related to MVB formation, protein sorting, and cargo transport.20 ESCRT-0 binds to ubiquitinated protein-specific endosomal membrane domains with the help of its ubiquitin-binding domain and thereby initiates the ESCRT mechanism. After this initiation, ESCRT-0 interacts with ESCRT-I and then with ESCRT-II, and the whole complex then connects to ESCRT-III, which ultimately helps promote vesicle budding. Then, the splitting of the buds occurs. A specific sorting protein, Vps4, is present to provide the energy that separates the ESCRT-III complex from the MVB membranes. TSG101 and CHMP4 are also linked to the generation of exosomes. Budding and secretion to the extracellular membrane are regulated by EXCRT protein complexes.21 However, there are also pieces of literature demonstrating ESCRT-independent pathways for cargo sorting. In 2013, Airola et al.22 revealed that raft-based microdomains in the plasma membrane help in the lateral segregation of cargoes in the endosomal membrane. Interestingly, these rafts are highly enriched with sphingomyelinases, which are essential enzymes for the formation of ceramide through the hydrolysis of phosphocholine. In these ceramide-dependent pathways, the lateral phase separation is induced by ceramide and also promotes the spontaneous formation of cone curvatures in the plasma membrane, aiding the budding process.23 The biogenesis of the exosome pathway is explained in Figure 2.
Figure 2.
Biogenesis of an exosome and its components. This image explains the ESCRT-dependent pathway and the ESCRT-independent pathway of exosome biogenesis in a more detailed manner and explains exosome-related structure components. Created with BioRender.com.
Regardless of the regulation of biogenesis, sorting, and budding, one chromaticism of the exosome is that it is comparatively smaller and more uniform in shape. This makes exosomes able to escape mononuclear phagocytes, reducing their circulation time and increasing cell-to-cell communication.24
3. Fundamentals of the Exosomes
3.1. Structure and Composition of Exosomes
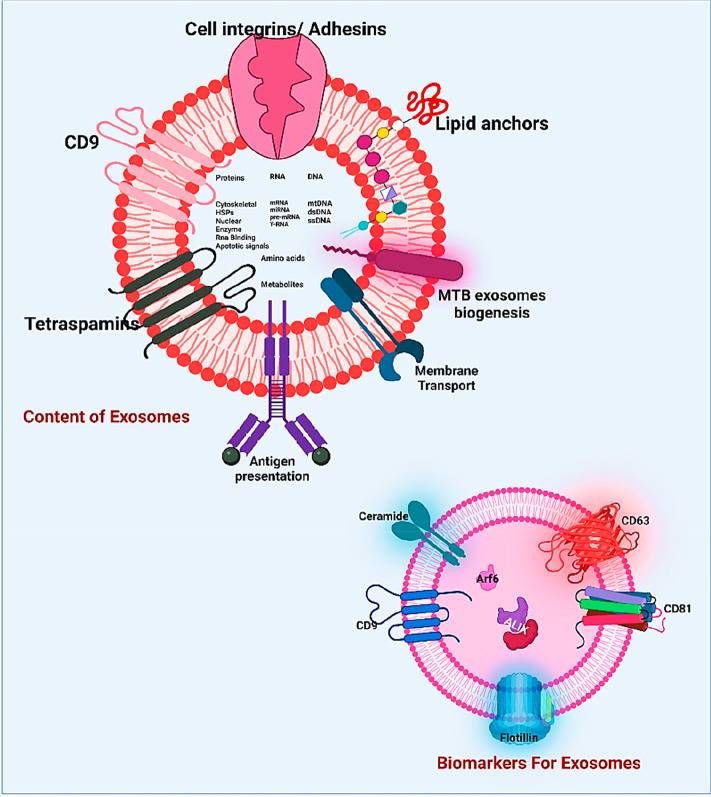
Exosomes construct a phospholipid outer envelope, and the inner core carries a group of biologically active molecules.3 Components of exosomes are proteins, lipids, nucleic acids, and glycoconjugates (Figure 3).
Figure 3.
Structure and composition of exosomes. Multiple exosome-associated components (protein, DNA, RNA, and surface marker) play vital roles in cancer biomarkers. Created with BioRender.com.
Exosome surface proteins play a principal role in cellular communication (such as integrins and tetraspanins). Tetraspanins mainly regulate cell communication facilitated by exosomes, and CD9, CD63, and CD81 are mainly observed. Not only tetraspanins but also many adhesin proteins help exosomes fix with the recipient cells.25 There are also reports showing the involvement of integrins in exosome-mediated metastasis. In 2015, Hoshino et al.26 showed the horizontal transmission of α6β4 and α6β1 to the lungs and the horizontal transmission of αvβ5 to the liver, which ultimately promoted metastasis to the respective organs. The main source of lipids in exosomes is the plasma membrane of the parent cell from which the exosomes originate, but apart from the plasma membrane exosomes can also be produced from Golgi membranes.27 Exosome membranes contain multiple lipid molecules, including ceramide, cholesterol, phosphatidylcholine, phosphatidylserine, phosphatidylinositol, sphingomyelin, phosphatidylglycerol, and many more.28 They also carry dynamic nucleic acids (mRNA, circRNA, tRNA, piRNA, tRNA, sncRNA, rRNA, lncRNA, mtDNA, and dsDNA).2,29−33
3.2. Exosome Sources
Exosomes are isolated from various biological fluids (blood, urine, saliva, etc.).34,35 The other sources of exosomes are from the tumor microenvironment, since a large number of exosomes are produced in tumors compared to normal cells. Apart from that, exosomes can come from HEK293 cells (human embryonic kidney cells), HeLa cells, and many more. Aside from being a source of cancer, exosomes are also a source of other notable substances. DCs, NKs, and exosomes released from tumor cells are the major sources of vaccine development. There have also been reports of exosome-based cancer vaccines made primarily from mesenchymal stem cells and macrophages.36,37
3.3. Exosome Isolation
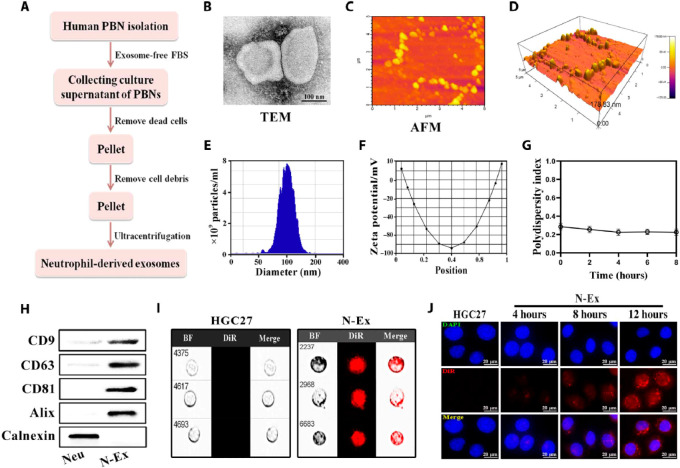
The isolation of exosomes is the most challenging process in EV research. There are several isolation methods, such as ultracentrifugation,38 density gradient centrifugation,39 ultrafiltration,40 size exclusion chromatography (SEC),41 immunoaffinity, and polymer precipitation.39 Each method has advantages and disadvantages. Some of the advanced techniques42 to isolate exosomes include microfluidics,42,43 lipid nanoprobes,44,45 and thermo-acoustofluidic separation.46,47 Exosome isolation is related to several methods, and their advantages and disadvantages are summarized in Table 1. In the experimental aspect, the appearance of an isolated exosome and exosome-specific biomarker analysis are explained in Figure 4.48
Table 1. Exosome Isolation Methods.
| isolation technique | mechanism | advantages | disadvantages | references |
|---|---|---|---|---|
| ultracentrifugation | components with varying sizes and densities have varying sediment speeds | a gold standard, ideal for large-scale samples, inexpensive, and the isolation procedure requires more than 4 h | exosomes may be damaged, the procedure is time-consuming and inconvenient, the purity is modest because of nonexosomal component contamination, and the yield is low | (38) |
| density gradient centrifugation | components with varying sizes and densities have varying sediment speeds | exosomal damage is avoided, hence we acquire high purity, and the procedure is completed in more than 16 h | preliminary preparation is labor-intensive, the procedure is time-consuming, and the yield is minimal | (39) |
| ultrafiltration | particles of varying sizes and molecular masses | it is simple and does not require any special equipment or reagents, it takes less than 4 h to complete, the component’s purity is high, and the yield is moderate | exosomes with tiny particle diameters are lost due to clogging on the filtering membrane | (40) |
| size-exclusion chromatography (SEC) | different sized and molecular density particles | exosome subtype isolation has a high level of specificity and it takes 0.3 h for qEV (Izon Science, New Zealand), which has a high yield and purity | lipoprotein contamination necessitates the use of special columns and packing | (41) |
| immunoaffinity | based on the interaction of antibodies with specific exosome membrane proteins | exosome subtype isolation has a high level of specificity, it takes between 4 and 20 h, has a high purity but limited yield | depending on the antibody’s specificity, it can be quite costly | (39) |
| polymer precipitation | the effect of exosomes on the solubility or dispersibility of high hydrophilic polymers | the simple technique takes between 0.3 and 12 h and is ideal for large-volume samples and the yield is high | contaminants may be present as a result of copurifying protein aggregates or residuary polymers, resulting in a low purity level. | (39) |
| microfluidics-based techniques | in this process, fluid runs through via a microchannel and captures the exosome based on a surface marker | this process supports isolated exosomes with high purity, reduced chemical utility and fast detection | expenses of this technique and it only applicable for small scale sample and have probability of losing exosome during washing time | (42, 43) |
| lipid nanoprobes | magnetic probe-mediated affinity-based exosome separation | its capable large-scale sample processing for protein and nucleic acids analysis | isolated exosome purity medium | (44, 45) |
| thermo-acoustofluidic separation | this process separated the exosome based on the lipid ratio | this process capable remove other extracellular vesicles contamination from exosome | protein contamination | (46, 47) |
Figure 4.
Neutrophil-derived exosome (N-Ex) isolation, characterization, and cell uptake. (A) Human peripheral blood neutrophil (PBN) isolation method. (B) Transamination electron microscopy analysis of N-Ex with 100 nm regulation. (C and D) Morphological analysis of N-Ex via Atom force microscopy (AFM). (E and F) Nanoparticle tracking assay (NTA) of N-Ex for size determination. (F and G) Explanation of the surface charge via ζ-potential analysis. (H) N-Ex surface marker analysis by Western blot with calnexin as the control. (I and J) Experimental analysis of DiR-labeled N-Ex untacking in the gastric cancer cell (HGC27) via (I) imaging flow cytometry and )J) fluorescence confocal laser microscopy. Cell nuclease staining was done by 4′,6-diamidino-2-phenylindole (DAPI) and bright field (BF) with 20 μm regulation. Reproduced with permission from ref (48). Copyright 2022 AAAS.
4. Exosomes for Cancer Theragnostic
4.1. Exosomes and Cancer
The association between exosomes and cancer is the most highlighted area of current research. The unexplained nature of exosomes has raised concerns about multiple events and their role in cancer cell angiogenesis and metastasis, including epithelial to mesenchymal transition (EMT) and immunological modulation.49 TEXs (tumor-derived exosomes) play an important role in the origin, development, and treatment resistance of cancer.50,159 The discovery of exosomes, which serve as regulatory agents in cancer intercellular communication, increases the potential to investigate the understanding of tumor immunity. Several scientific studies suggest that tumor-associated macrophages (TAMs) are involved in major inflammation, suggesting that TAMs play a significant role in tumorigenesis.51 According to various studies, TAMs promoted multiple cells in macrophage polarization.52 TAMs lose their anticancer activity and promote tumor progression. Exosomes released from the tumor reprogram the macrophages and support cancer development.53 Hypoxia is another important feature of the TME (tumor microenvironment) related to immunosuppression. Hypoxic conditions influenced the tumor-cell-derived exosome to drive cancer to a more aggressive pattern.54 The epithelial to mesenchymal transition (EMT) can be regulated by several transcription factors.24 Hypoxic tumor cells, derived from multiple molecules of exosomes, reprogram the immune system and promote cancer development. This exosome miRNA cargo affects macrophage function and M2 polarization.55 Exosomes are associated with multiple miRNAs associated with tumor progression.56,57 The exosome circular RNAs play a crucial role in the cellular communication that occurs in the tumor microenvironment. In addition to RNA, proteins also play a crucial role in tumor progression. Matrix metalloproteinases (MMPs) are related to cells with cellular adhesion properties, and TEXs alter MMP functions, causing cells to become motile. It was discovered that M2 macrophage-derived exosome CD11b/CD18, an integrin, promotes cancer cell proliferation while inhibiting metastasis by activating MMP-9. Due to its antiatherogenic effects, apolipoprotein E (ApoE) is an important protein molecule involved in M2 polarization. TAM releases IL-1, VEGF (vascular endothelial growth factor), and cytokines that participate in tumor development. Exosomes, which carry multiple cargoes to accelerate angiogenesis, were recently discovered to play a critical role in cancer invasiveness.53 In cancers, TEXs are also responsible for theepithelial to mesenchymal transition (EMT).58 The surface integrin of exosomes leads to organ-specific metastasis. TEXs-guided cancer cell migration in a specific organ is regulated via the diversity of TEX integration.26 The transcriptional regulator GATA3 was abundantly released from TAM-derived exosomes, where it plays an important role in epigenetic modulation to induce angiogenesis and EMT.53
4.2. Exosome is the Source of Cancer Biomarkers
The molecular contents of exosomes normally reflect those of their parent cells and can therefore be used as biomarkers for pathophysiological complications (such as cancer).59,35,158 Tumor and stromal cells in the TME have been reported to release exosomes, and their molecular signatures play a dynamic role in cancer.60,61 For example, it has been found that TNBC (triple-negative breast cancer) cells with CCL5 on their surfaces, derived from tumor-derived exosomes, alter TME-associated macrophages and develop a metastatic nature, resulting in a TME favorable for carcinogenesis.62 Researchers suggest that derived cancer stem cells are involved cancer metastesis.63 TEXs are being studied as diagnostic and prognostic biomarkers in clinical trials. A clinical study NCT04523389 related to colon cancer focuses on the development of diagnostic markers. TNBC TEVs carry multiple molecules that are sources of diagnostic and prognostic biomarkers.60 Some of the most complicated cancers, such as breast cancer,64,69 lung cancer,65,70 colon cancer,66,71 prostate cancers,67,72 and liver cancers,68,73 and their related exosome biomarkers with clinical importance are discussed in Table 2.
Table 2. Exosome-Associated Cancer Biomarkers and Their Clinical Significance.
| biomarker | cancer | source | exosome component | clinical significance | reference |
|---|---|---|---|---|---|
| diagnostic | breast cancer | plasma | miR-223-3p | early diagnostic breast metastasis biomarker | (64) |
| lung cancer | serum | miR-106b | it is highly expressed in serum and it is also associated with lymph node metastasis and mmp protein expiration in lung cancer metastasis | (65) | |
| colon cancer | plasma | CD147 | it highly expresses in colon cancer patients | (66) | |
| prostate cancers | urine exosome | miRNA-501-3p | it is downregulated in prostate cancers but suppresses E-cadherin expression and promotes metastasis | (67) | |
| liver cancers | serum | circRNA-100338 | it enhances liver cancer metastasis | (68) | |
| prognostic | breast cancer | plasma | miR-222 | it is interlinked in breast cancer (highly expressed) with lymphatic metastasis | (69) |
| lung cancer | plasma | miR-451a, | it participates in lymph node metastasis in lung cancer | (70) | |
| colon cancer | serum | miRNA-203 | it highly expressed colon cancer and is associated with metastasis, in vivo model (liver metastasis) | (71) | |
| prostate cancers | plasma | miR-1290 and miR-375 | it highly expressed prostate cancer and is related to castration-resistant poor overall survival | (72) | |
| liver cancers | serum | miR-1262 | it is an efficient prognostic biomarker of liver cancer | (73) |
4.3. Exosomes as Carriers
Exosomes are nanosized extracellular vesicles released by multiple cells. Exosomes with a wide size distribution are easier to internalize, as cells prefer smaller exosomes.64 Because of their economy of scale and immense potential in drug therapy, they have been an important research area in biomedicine and biomaterials.65 Exosomes are released into the surrounding body fluids. They have been shown to contain the molecular signatures of the parent cells (such as proteins, DNA, RNA, and lipids). This signature molecule acts as a messenger of cell status. Exosomes are the most interesting noninvasive diagnostic biomarkers and therapeutics. Their cargo molecules are involved in cellular communication.24,60 The secretion of exosomes from specific cells or tissues is based entirely on the cellular and philological condensation of cells.66 The exosome leads to biologically active molecules.67 The exosomal molecular signature has a complex association with multiple treatment resistance and carcinogenesis.60 miRNAs associated with TEXs promote EMT (miRNA-21, miRNA-92b, miRNA-130a, miR-149, miRNA-181c, miRNA-200, miRNA-328, miRNA-423-5p, miRNA-602, and miRNA-1246), tumorigenesis, invasion, and metastasis (let-7a miRNA, miRNA-21, miRNA-221/222, and miRNA-42.68
4.4. Routes of Administration
Understanding and comprehensively analyzing the underlying complexity of cellular communication is a potential tool for the development of efficient drug delivery systems and therapies in the fight against cancer. In the past decade, significant research in the field of exosomes has gained momentum. The whole situation regarding their cellular interactions with disease progression has yet to be fully explored.69 Recent scientific expeditions have documented effective exosome-mediated therapeutic delivery to cancer models and provided insights to improve disease pathophysiology.70 Efficient drug loading and sustained drug release via exosomes in and around the tumorigenic tissue depends on a complex, multifaceted set of factors.71 Based on clinical data and other medical research, there are specific drug delivery routes that exosomes should follow in order to reach the tumor target site.71,42 Nowadays, several conventional and unconventional routes of administration for these vesicles have been tried by several clinical research groups, namely, parenteral, oral, intertumoral, intranasal, and intraperitoneal routes.72−74 Needless to say, the appropriate choice of the route of administration of the drug in relation to the type of cancer it is dealing with is absolutely essential to the success of exosome delivery. Considering all the challenges and adversities, these exosome-based drug delivery targets pave a new way toward successful drug delivery and sustained drug release strategies in various tumors.74 Appropriate clinical trials and research need to be standardized to target potential exosomal agents to combat the growing rates of cancer.75,76
4.5. Exosome Loading Method
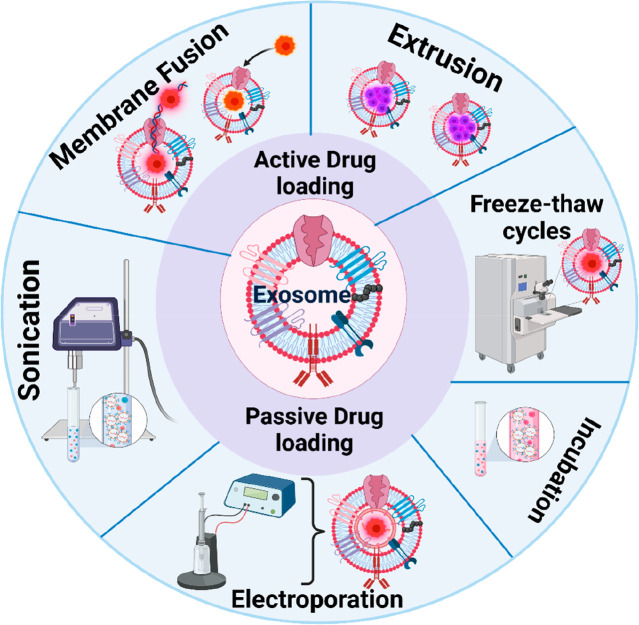
Exosomes are natural carriers into which drugs can be loaded. Exosomes are encapsulated with drugs to make them suitable for the various target therapies. There are three different methods by which drug encapsulation occurs: the postloading method, the preloading method, and the fusion method.77 Therefore, since the incorporation of the drug into this lipid bilayer membrane is challenging,78 two different methods are followed; active loading and passive methods79 (Figure 5).
Figure 5.
Exosome drug loading methods. Exosome drug loading methods are classified into two major classes: active (electroporation, sonication, fusion method, freeze–thaw cycles, and use with membrane permeabilizers) and passive (incubation). Created with BioRender.com.
In active or remote or postdrug loading, the cells are cleaned to obtain a naïve exosome that is then sealed with drugs, while in passive loading or preloading methods the cells and the drugs are incubated together and the component later undergoes purification to yield a drug-sealed exosome. The postloading method works better with hydrophobic drug components than hydrophilic drug components.80
5. Active Drug Loading Approaches
Rupture of the exosome membrane is used to allow the entry of functional components into the exosome during drug loading. After the required molecules are loaded into the exosomes, the exosome retains its previous shape. Electroporation sonication, extrusion, and freeze–thaw cycling are some of the methods used to disrupt exosome membranes.77 Studies suggest that the active drug loading method increases the drug encapsulation efficiency of exosome development 11-fold.81 The limitation of this method is that it can affect exosome targeting properties and the native structure during the membrane rupture process.77
5.1. Electroporation
Electroporation involves a high-intensity electric field, instantaneous changes in cell membrane permeability, and drug loading. The voltage settings for different types of donor cells, such as B. Hela cells, monocytes, and immature dendritic cells, generally range from 150 to 700 V.77 Drug molecules enter through holes created in the exosome membrane during electroporation, while the membrane is restored after loading. This approach is commonly used to load large molecules such as miRNAs and siRNAs81 into exosomes. The electroporation process has a poor loading capacity due to the aggregation of RNA and stability issues. This approach can improve the loading of hydrophilic small molecules in exosomes and increase the efficiency of RNAs in exosomes.81
5.2. Sonication
The premise of ultrasonic drug loading is that ultrasonic waves lower the microviscosity of the membrane (usually by at least twofold), allowing the hydrophobic drug to pass.77 Exosomes derived from parental cells or recipient cells are mixed with a specific drug and protein legend before being sonicated with a homogenizer probe. The integrity of the exosome membrane is disrupted by the mechanical shear stress generated during sonication, allowing bioactive chemicals to enter the exosome while the membrane is deformed.80 Research suggests that sonication alters the viscosity of exosomes,68 but there are no reports of a reduction in the membrane-bound protein or lipid content of the exosome.80 After a 1 h incubation at 37 °C, it was shown that the membrane integrity of the exosome was restored. Drugs that bind to the surfaces of exosomes release very quickly, and drugs encapsulated via the exosome take time to release phage.82
5.3. Fusion Method
Membrane fusion, itself a scientific achievement, can fuse exosomes and nanocomposites within a membrane structure. It allows for the prolonged release of nanodrugs, enhances absorption and efficacy, and performs an exocrine function in immune system response, antigen presentation, cell migration, cell differentiation, and tumor invasion.77 This adaptable technique was successful in enriching exosomes using hydrophilic biological components without removing their function. When a drug and a liposome-encapsulated drug were compared, hybrid EVs increased the cellular transport efficiency of a chemotherapeutic agent by three- to fourfold. Fluorescence resonance energy transfer, which detects changes in nanoscale spacing of biological macromolecules in vivo, was used to confirm the hybrids.83
5.4. Freeze–thaw Cycles
Exosomes are incubated with selected drugs at room temperature for a set period before being quickly frozen at −80 °C or in liquid nitrogen. Thereafter, the combination is allowed to thaw at room temperature. Freeze–thaw cycles are performed at least three times to improve drug encapsulation. Compared to sonication or extrusion, this method has a reduced drug loading capacity. Furthermore, this approach can increase exosome aggregation, resulting in large-scale drug loading of exosomes.84,85,83
5.5. Used with Membrane Permeabilizers
Membrane permeabilizers and surfactants such as saponin can interact with cholesterol in the cell membrane to create pores that allow the passage of exosomes. The membrane permeability approach can improve the loading efficiency of catalase into exosomes compared to the incubation method.80
6. Passive Loading Approach
The method involves the integration of drugs with exosomes. The mechanism of encapsulation and its loading efficiency depend on the hydrophobic interaction and diffusion between the loaded molecule and the lipid layer of the exosomes.85,77
6.1. Incubation
The passive loading approach involves two different types of incubation: incubation of drug along with an exosome or with donor cells. In the case of incubating a drug with exosomes, this technique allows the drug to enter the exosome based on the concentration gradient during the incubation. Since hydrophobic drugs can interact with the lipid surfaces of exosomes, this property is exploited for drug loading.80 In one study, exosomes were incubated with the paclitaxel stock solution for 1 h at 22 °C to produce an excipient preparation with a loading efficiency of 9.2%. Based on the high lipophilicity and limited water solubility of paclitaxel, this technique uses the passive diffusion of drugs packaged in exosomes. In addition, it has been suggested that coincubation at 37 °C can be used to load miRNAs into exosomes.77 The disadvantages of this method is that it is limited to a specific type of drug and the amount released after incubation is not sufficient for clinical trials.86 In the case of incubation with donor cells, the drug is coincubated with the donor cells, which is done by pretreating the cell membrane, and then exosomes loaded with the drug are shed using UV light, heat, or both. In both cases, the cell membrane is unobstructed, but the downside that researchers face during incubation is the insufficient number of exosomes that are secreted.13 The efficiency during loading and the cytotoxicity that cells experience while responding to the drug also pose research challenges.80
6.2. Drug Delivery via Exosomes
The latest discoveries point to a unique property of exosomes, as it was found that exosomes can transport proteins and genetic and epigenetic information from one cell to another cell through receptor–ligand interactions.87,88 One of the results suggests that exosomes obtained from mouse mastocytes can be transferred to humans and the RNA obtained from this transfer can be used in other humans and mice.2 Discoveries stated that exosomes self-decode upon transfer into recipient cells according to the host body, hence protein translation in the host body occurs depending on the host physiology.89 The uniqueness of the exosome makes it the most important medium for transporting drugs to the cells.48 Unlike other carriers used in cells, such as liposomes and polymeric nanoparticles, exosomes have the unique potential of being an endogenous cellular machinery that can be used for drug delivery and storage.90 Exosome delivery enables simultaneous intercellular communication by sending many signals simultaneously. Exosomes are unlikely to be freely circulating soluble factors and can release large amounts of functional molecules, as they are soluble factors in the host cells.91 Exosomes have other additional properties such as the protection of the protein or drug entrapped within due to their small size, which helps exosomes avoid phagocytosis.13 Exosome cargoes can travel long distances, have high biocompatibility, are nonimmunogenic and targeted, and can overcome a variety of physical barriers due to their properties.13,92
6.3. Delivering Small Molecules via Exosomes
Drugs can be encapsulated in exosomes, thereby prolonging the drug half-life and improving the stability of drug release. Furthermore, due to their endogenous origin, exosomes are highly biocompatible and can be used as nanocarriers for tissue-specific targeted delivery.86 Studies show that exosomes were designed with hydrophobic agents such as curcumin, and the results showed that exosomes could carry the hydrophobic agent and also enhanced its anti-inflammatory properties.93 Various studies conducted have found that exosomes can cross the blood–brain barrier. This scientific evidence suggests that exosomes overcome nanoparticles based on multiple membrane cross-constraint. This attribute of the exosome makes it a more efficient drug delivery tool.94 From this we can conclude that exosomes can not only transport the drugs but also increase their half-life, reduce toxicity, and even overcome various barriers.
6.3.1. Delivering Proteins via Exosomes
Exosomes are also used to carry large molecules, such as proteins, in addition to tiny compounds. To understand the role and importance of exosomes in protein delivery, we can consider a case related to Parkinson’s disease (PD).13 Exosomes produced by the central nervous system (CNS) have been found in cerebrospinal fluid and peripheral body fluids, and several studies suggest that their molecular signatures play a role as biomarkers in Parkinson’s disease (PD). Exosomes have been shown to spread toxic α-synuclein protein (syn) between cells and cause apoptosis, suggesting a critical mechanism that causes the disease. This accelerates syn-aggregate proliferation in brain pathogenesis in Parkinson’s disease. However, exosomes have also been reported to play a significant role in the treatment of PD. In the mouse model of PD, researchers have found that exosomes transport catalase and small interfering RNAs to the brain.95 Designing exosomes with catalase can be said to be a promising therapy for PD therapy because the delivery of catalase across the BBB, like many other drugs, is challenging and exosomes have overcome this hurdle. The targeted delivery of armed exosomes is also used as an anticancer treatment, with the exosomes loaded with various active pharmaceutical ingredients (API), including genetic material, proteins, and chemotherapeutic agents.96 The exosomes have a more efficient ability to load anticancer drugs onto their surfaces compared to synthetic nanoparticles.
6.3.2. Delivering Genetic Material via Exosomes
Various studies conducted have found that exosomes can carry both large and small molecules. These cargoes can be engineered to even carry genetic and epigenetic material.87 Gene therapy is being considered for the treatment of various types of cancer. Exosome-based gene therapy transports siRNA, mRNA, and miRNA along with exosomes.97 Exosomes are the most efficient miRNAs transporter tools and are used for therapeutic RNA delivery.98 Several studies show that exosomes transport RNA more efficiently than any other nanoparticle. Exosome-based small RNA delivery enhances its functional efficiency. Studies have shown that exosomal miRNAs molecules have a complex interrelationship in multiple cancer delivery phages (angiogenesis and metastasis).97
7. Application of Exosome-Based Drug Delivery in Multiple Cancers
Exosomes are nanosized extracellular vesicles. They secretes from almost all cells. The main contribution of exomes is in cellular communication.99,100 They have been found in body fluids such as blood, urine, cerebrospinal fluid, saliva, etc. This evidence proved that they are involved in several physiological metabolic processes.100 However, exosomes have also been shown to be involved in cancer development, progression, and metastasis. Tumor-derived exosomes (TDXs) have been reported to promote cancer proliferation and cause the formation of the premetastatic niche. They have also been found to regulate drug resistance.13,100 TDXs turn the recipient cells into cancer cells. Evidence has shown their involvement in the modulation of immune response, stromal cell reprogramming, extracellular matrix remodelling, the induction of drug resistance, etc.101 Exosome-associated molecular signatures are promising evidence for the invention of cancer biomarkers.26 The exosome-based therapeutic approach is the most innovative area in cancer research.13,100 This Review aims to summarize the clinical therapeutic exosomes that behave as nanocarriers that deliver nucleic acids, mRNAs, microRNAs, proteins, lipids, and metabolites to other cellular habitats and behave as convenient drug delivery systems.25 The exosomes are isolated from the patients and conjugated with drugs, and this approach develops biocompatibility and low toxicity in drug delivery.102 This system also bypasses the P-glycoprotein drug efflux system, thus reducing the risk of drug resistance.82 It has been reported by a research group that the exosome penetrates deep into the tissue, effectively diffuses in the blood, and even crosses the biological barrier.103 Exosomes can also be effectively engineered for cell and tissue specificity, allowing the increase of the drug concentration at a given diseased site.104 The potential applications of exosome-based cancer therapy are presented in Table 3. Homeostasis in a normal cell is maintained by the transfer of bioactive molecules across membranes. This diffusion and uptake of biological materials occurs through extracellular vesicles, which characterize the cargo and send it to its assigned destination. Exosomes are extracellular vesicles that moderate this intercellular communication. Previous studies have shown that exosome cargoes can hijack the cells in several pathological conditions such as cancer.13,100 Therefore, they have emerged as the essential regulatory molecules that modulate cell-to-cell communication during phage. The exosome has been shown to have an important interaction between tumor chemotherapeutic resistance and cancer metastasis.105 In the recent past, therefore, exosomes have been considered as important diagnostic biomarker sources and therapeutic tools against cancer. Although exosomes have shown promising results in vitro and in vivo, their use in humans as cancer therapeutics is still under investigation. Exosomes require more detailed study and understanding to become potential drug delivery systems and anticancer therapies in the near future.
Table 3. Exosomes for Targeted Drug Delivery in Cancer Therapy.
| therapeutic cargo | targeting ligand | target cell | function | method of synthesis | types of modification | reference |
|---|---|---|---|---|---|---|
| KRAS siRNA (Kirsten Ras oncogene short interfering RNA) | iRGD peptide (Arg-Gly-Asp peptide) | adenocarcinoma, human alveolar basal epithelial cells | targets oncogenic KRAS (Kirsten Ras oncogene) | LAMP-2B (Lysosome-associated membrane protein 2 gene) | genetically modified | (105) |
| DOX (doxirubin) | αv-integrin-specific iRGD peptide | breast cancer | targeted delivery of DOX (doxirubin) | LAMP-2B | genetically modified | (105) |
| SOX2 siRNA (silencing RNA) | tLyp-1 (linear truncated form of LyP-1) | nonsmall cell lung cancer, A549 stem cells | Gene delivery for cancer therapy | LAMP-2B | genetically modified | (106) |
| imatinib, BCR-ABL siRNA | IL-3 | chronic myelogenous leukemia cells | inhibits cancer cell growth, increased intratumoral accumulation | LAMP-2B | genetically modified | (107) |
| 5-fluorouracil anti-miRNA-21 | zHER affibody | colorectal cancer | Reverses chemoresistance and improves cancer treatment efficiency | LAMP-2B | genetically modified | (108) |
| Tpd50 siRNA | DARPin | HER2-positive cells | RNAi therapy of HER2-positive cancer | LAMP-2B | genetically modified | (109) |
| miRNA-let7a | GE11 peptide | breast cancer | targets EGFR-expressing tumors | LAMP-2B | genetically modified | (110) |
| Smart-exos | αCD3/αEGFR | T cells (Jurkat), EGFR-positive breast cancer | cell-free cancer immunotherapy | Smart-exos | genetically modified | (111) |
| miRNA-26a | ApoA-1 | hepatocellular carcinoma (HepG2) | suppresses tumor cell migration and proliferation | CD63 | genetically modified | (112) |
| antigen | OVA antigen | CD8+ T cells | improves the immunogenicity of cancer vaccines | CD63 | genetically modified | (113)(114), |
| Sstreptavidin-HRP, mannosamine | l-azidohomoalanine (AHA) (azide-bearing amino acids) and saccharides | biotin receptors | Florescence of cancer cells | Exosome azide integration, DBCO-PEG4-biotin–avidin conjugation | chemically modified | (115) |
| curcumin-SPION | neuropilin-1-targeted peptide | glioma | simultaneous diagnosis and treatment of glioma | click chemistry | chemically modified | (116) |
| paclitaxel (PTX) | AA | murine lung cancer, sigma receptor-positive cells | improves drug circulation and inhibits pulmonary metastases | DSPE-PEG-AA | chemically modified | (117) |
| quantum dot photothermal agent | RGD | breast cancer | near-infrared-II region quantum dot delivery for nucleus-targeted low-temperature photothermal therapy | DSPE-PEG-RGD | chemically modified | (118) |
| elastin | folate | breast cancer | targeted induction of ferroptosis | DSPE-PEG-folate | chemically modified | (119) |
| surviving siRNA | PSMA RNA aptamer, EGFR RNA aptamer, folate | breast cancer, prostate cancer, colorectal cancer | tumor-targeted RNAi nanomedicine | chol | chemically modified | (119) |
| miRNA-let7, VEGF siRNA | AS1411 aptamer | nucleolin-positive cancer cells | tumor-targeted small RNA delivery | chol | chemically modified | (119) |
| DOX | sgc8 aptamer | leukemia cells | targeted anticancer therapy | diacyl lipid-(PEG)2 | chemically modified | (120) |
| PTX | AS1411 (aptamer-conjugated) | breast cancer | targeted anticancer chemotherapy | chol-PEG2000 | chemically modified | (121) |
| methotrexate, KLA (Lys-Leu-Ala) | ApoA-1 mimetic peptide | glioma | selective brain tumor treatment (glioblastoma multiforme) | lipid | chemically modified | (122) |
| photosensitizer | NLS peptide | carcinoma (4T1), colorectal cancer (CT26) | dual-stage light-guided plasma membrane and nucleus-targeted photodynamic therapy | C16 | chemically modified | (123) |
| aSIRPα, aCD47 | antibodies | macrophages and tumor cells | enhance phagocytosis of cancer cells by blocking SIRPα-CD47 interaction | azide-modified | chemically modified | (124) |
| mannosamine | RGD | αvβ3 -overexpressing cells (HUVEC) | promote angiogenesis with targeted imaging | DSPE-PEG-RGD | chemically modified | (125) |
| SIRPα | mRNA | embryonic fibroblasts (MEFs) | increased exosome circulation time | CD47 surface decoration | chemically modified | (126) |
| copper-64 (64Cu)-radiolabeled polyethylene glycol (PEG) | N/A | diagnosis of cancer (passive action) | reduced exosome clearance enhanced tumor penetration | surface PEGylation | chemically modified | (127) |
8. Clinical Applications of Exosomes in the Treatment of Cancer
8.1. Advancements and Limitations
Exosomes have great potential as new drug delivery vehicles due to their inherent involvement in intercellular exchange of biomolecules, particularly for biotherapeutics that can be loaded into exosomes using the cellular EV packaging machinery.90 As we discussed earlier, delivering a drug to target sites and crossing the barriers was possible through exosomes compared to other nanoparticles. The research data reported that the drug potency and half-life of exosomes were well maintained when they were introduced into the recipient cell. Since the exosome is a natural mediator, it has the natural ability of cell permeability, which helps it cross physical barriers and even escape lysosomal degradation and the endosomal pathway.13 Macrophage-derived genetically engineered exosomes are capable of drug delivery without rejection.128
There are several underlying questions that remain unanswered that limit the use of this novel component.129 (1) Industrial-scale production of exosomes would help treat cancer. (2) The storage of these exosomes derived from different cells and their longevity when not in use. (3) Targeting the armed exosomes to perform biogenesis at the site and not with other exosomes already present in the recipient cells. (4) The pathways and mechanisms that control exosomes will eventually help researchers fully control drug-containing exosomes. (5) One way to prevent therapeutic exosomes from reacting with healthy cells is to evaluate the characteristics of pharmacokinetics and pharmacodynamics, as well as safety, feasibility, toxicity, and pharmacodynamics. (6) Although exosomes are the natural mediator of cells, the immune response of a loaded exosome in the body has yet to be discovered.13 (7) We lack a technique that can help us to isolate exosomes with high purity and in reasonable quantities, which could help us to reduce costs, since exosome isolation is very expensive.13 (8) Hybrid exosomes are being used based on future demand, but the chemical efficacy and safety of such exosomes have yet to be investigated.13 (9) Exosomes are composed of heterogeneous components and have been reported to play an important role in tumor growth and even metastasis. Therefore, the immunogenic response of the hybrid exosomes or exosomes derived from other animals must be thoroughly investigated before they are used for clinical trials.13 (10) Although experiments show promising results in removing components from macrophage-derived exosomes by hypotonic treatment,128 the effect of the same treatment on exosomes bearing caspase-3 or other carcinogenic components remains to be investigated.39 (11) Most anticancer exosome drugs are still in the early stages of development.130
8.2. Exosome-Based Drug Delivery-Associated Clinical Trial for Cancer
Exosome-based clinical trials related to drug delivery are the most highlighted research area today. Sometimes they use a combination of traditional cancer therapy to develop effectiveness. Multiple cancer types and associated clinical trials of exosome-based drug delivery are constructively summarized in Table 4.
Table 4. Clinical Trials of Exosome-Associated Drug Delivery in Multiple Cancers.
| cancer type | drug used | clinical trial ID | sponsor | reference |
|---|---|---|---|---|
| breast cancer, Her-2 positive | trastuzumab emtansine | NCT01772472 | funded by F. Hoffmann-La Roche/Genentech, KATHERINE ClinicalTrials.gov number NCT01772472. | (131) |
| breast cancer, triple-negative | atezolizumab with chemotherapy | NCT02425891 | F. Hoffmann-La Roche/Genentech, IMpassion130 ClinicalTrials.gov number NCT02425891 | (132) |
| breast cancer, Her-2 negative | ribociclib with endocrine therapy | NCT02278120 | Novartis, MONALEESA-7 ClinicalTrials.gov number NCT02278120. | (133) |
| lung cancer | immunotherapy atezolizumab in combination with chemotherapy | NCT02763579 | F. Hoffmann-La Roche/Genentech, IMpower133 ClinicalTrials.gov number NCT02763579. | (134) |
| colorectal cancer | encorafenib (BRAF inhibitor) plus cetuximab or encorafenib plus cetuximab and binimetinib | NCT02928224 | Amgen (Inst), Bayer (Inst), Boehringer Ingelheim (Inst), Eli Lilly (Inst), Novartis (Inst), Roche (Inst), Celgene (Inst), Ipsen (Inst), Merck (Inst), Merck KGaA (Inst), Servier (Inst), Bristol-Myers Squibb (Inst). Genentech (Inst), Bayer (Inst), Pfizer (Inst), Eisai (Inst), Eli Lilly (Inst), Boston Biomedical (Inst), Daiichi Sankyo (Inst), Array BioPharma (Inst), Array BioPharma, GlaxoSmithKline, Novartis, Merck Serono | (135) |
| prostate cancer | combination of enzalutamide with androgen suppressor | NCT02446405 | Astellas Scientific and Medical Affairs and others; ENZAMET (ANZUP 1304) ANZCTR number ACTRN12614000110684; ClinicalTrials.gov number NCT02446405, and EU Clinical Trials Register number 2014-003190-42 | (136) |
| renal cell carcinoma | axitinib and pembrolizumab | NCT02853331 | Merck Sharp and Dohme, KEYNOTE-426 ClinicalTrials.gov number NCT02853331. | (137), (138) |
| combination of avelumab and axitinib | NCT02684006 | Pfizer and Merck (Darmstadt, Germany), JAVELIN Renal 101 ClinicalTrials.gov number NCT02684006 | ||
| brain cancer | temozolomide with radiation selumetinib | NCT01149109 | German Federal Ministry of Education and Research | (139), (140) |
| NCT01089101 | National Cancer Institute Cancer Therapy Evaluation Program, the American Lebanese Syrian Associated Charities, AstraZeneca | |||
| lymphoma | rituximab with or without lenalidomide | NCT01938001 | Celgene Corporation (Summit, NJ) | (141) |
| leukemia | c-methotrexate or high-dose methotrexate | NCT00408005 | National Cancer Institute (NCI) | (142) |
| hepatoblastoma | minimal adjuvant chemotherapy | NCT00980460 | National Institutes of Health. | (143) |
| melanoma | dabrafenib and trametinib | NCT01972347 | GlaxoSmithKline; Novartis; National Health and Medical Research Council, Australia; Melanoma Institute, Australia. | (144), (145) |
| ipilimumab and nivolumab | NCT02977052 | Bristol-Myers Squibb |
9. Future Perspectives
Despite promising experimental achievements, there are some challenges in exosome-based drug delivery in terms of heterogeneity in origin, structure, and function. Among all these limitations, the greatest concern is the nonspecificity of exosome biodistribution. They can be found in various bodily fluids in the human body.146 However, in a study on BALB/c nude mice, it was observed that in the case of pancreatic cancer exosomes secreted by Panc-1 cells accumulate at the site of the tumor in a time-dependent manner. The rate of exosome accumulation is 30× higher than that of PEG–PE micelles at 4 h postinjection.147 Another major problem of exosomes is their ability to be rapidly cleared from the bloodstream after in vivo administration.148 This property is mysterious, since the exosome itself is made up of unique protein–lipid assemblies. However, the mystery was solved in a study that found the rapid clearance of exosomes from the bloodstream is due to uptake by macrophages. Experimental results clearly showed that exosomes derived from B16–B16 cells are quickly cleared after intravenous injection because liver and spleen macrophages have captured them.149 This problem can be solved to some extent by incorporating polyethylene glycol (PEG) into the structures of exosomes. It has been experimentally confirmed that exosomes with PEG can be detected even after 60 min postinjection, while exosomes without PEG can only be detected for 10 min.150 The implication of exosomes as drug carriers for unconventional therapeutics,151 including ocular, pulmonary,152 cutaneous, etc., is also difficult. To improve this, many parameters came into play. Two of the most important things are the penetrating power of exosomes in different tissues, tight junctions, etc. and their ability to evade the attack of tissue-resident immune cells and enzymes.153 The low yield of exosomes is a concern, as less than 1 g of protein is produced per ml of cell culture.154 Therefore, in order to conduct an experiment or clinical study, a large number of cells must be cultured. This limitation can be managed using exosome-mimetic nanovesicles.155 Exosome-mimetic nanovesicles (EMNV) can be produced by the serial filtration of extruded cells.154 It is reported that in this way the yield can be increased up to 100-fold.155 Plant-derived exosomes are some of the most frequent directions for research in the future. It has been reported that there are some exosome-related nanoparticles called folic acid-modified ginger-derived nanovectors that show very high compatibility and high potency while targeting cancer cells.156 In the case of FDA-approved nanomedicine research, the primacy of the exosome is limited. There are several aspects, including selecting the source of exosomes, standardizing techniques for culturing cells that produce exosomes, and isolating and quality controlling produced exosomes so that they can be applied to health-related problems, with particular reference to cancer. New technologies and regulations could reduce the boundaries of these fields.153 Finally, exosome-based research requires interdisciplinary155,156 work ecosystems that can develop an exosome-based advance therapeutic tool (such as a cancer vaccine157) for future cancer-associated global health problems.
10. Conclusions
Exosomes are burgeoning as next-generation platforms for nanomedicine in cancer therapy. It is clear that exosomes are used as promising biomarkers for several potential cancer types and also as an early detection tool in many clinical studies, some of which have already been discussed in this Review. The biocompatibility of exosomes and their highly specific interactions in living systems have stimulated the development of futuristic exosome-based therapeutic and drug delivery approaches. Genetically engineered exosomes loaded with specific drugs that target specific cancer cells offer more benefits compared to traditional cancer therapies. Nonetheless, this innovative approach also has some limitations in terms of difficulties in its scalability, purity, and isolation methods. This is the area where deeper research is needed. In the future, more efforts and more investigations will contribute to the development of this field, which will definitely open a new door through which we can be one step ahead of personalized medicine to treat cancer.
Acknowledgments
N.D.T. acknowledges funding under the Science Foundation Ireland and Irish Research Council (SFI-IRC) pathway program (21/PATH-S/9634).
Author Contributions
∇ Equally contributing authors.
The authors declare no competing financial interest.
References
- Harding C. V.; Heuser J. E.; Stahl P. D. Exosomes: looking back three decades and into the future. J. Cell Biol. 2013, 200 (4), 367–71. 10.1083/jcb.201212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H.; Ekstrom K.; Bossios A.; Sjostrand M.; Lee J. J.; Lotvall J. O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9 (6), 654–9. 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Raposo G.; Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200 (4), 373–83. 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaj L.; Lessard R.; Dai L.; Cho Y. J.; Pomeroy S. L.; Breakefield X. O.; Skog J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J.; Würdinger T.; van Rijn S.; Meijer D. H.; Gainche L.; Curry W. T.; Carter B. S.; Krichevsky A. M.; Breakefield X. O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10 (12), 1470–1476. 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graner M. W.; Alzate O.; Dechkovskaia A. M.; Keene J. D.; Sampson J. H.; Mitchell D. A.; Bigner D. D. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009, 23 (5), 1541–57. 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. J.; Lim J. W.; Moritz R. L.; Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev. Proteomics 2009, 6 (3), 267–83. 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- Mathivanan S.; Fahner C. J.; Reid G. E.; Simpson R. J. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012, 40 (D1), D1241–D1244. 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside T. L. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv. Clin Chem. 2016, 74, 103–41. 10.1016/bs.acc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchorska W. M.; Lach M. S. The role of exosomes in tumor progression and metastasis (Review). Oncol. Rep. 2016, 35 (3), 1237–44. 10.3892/or.2015.4507. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Cao X. Organotropic metastasis: role of tumor exosomes. Cell Res. 2016, 26 (2), 149–50. 10.1038/cr.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.; Zhong J.; Zhong B.; Huang J.; Jiang L.; Jiang Y.; Yuan J.; Sun J.; Dai L.; Yang C.; Li Z.; Wang J.; Zhong T. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett. 2020, 476, 13–22. 10.1016/j.canlet.2020.01.033. [DOI] [PubMed] [Google Scholar]
- Ha D.; Yang N.; Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm. Sin B 2016, 6 (4), 287–96. 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai R. C.; Yeo R. W.; Tan K. H.; Lim S. K. Exosomes for drug delivery - a novel application for the mesenchymal stem cell. Biotechnol Adv. 2013, 31 (5), 543–51. 10.1016/j.biotechadv.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Aqil F.; Munagala R.; Jeyabalan J.; Agrawal A. K.; Kyakulaga A. H.; Wilcher S. A.; Gupta R. C. Milk exosomes - Natural nanoparticles for siRNA delivery. Cancer Lett. 2019, 449, 186–195. 10.1016/j.canlet.2019.02.011. [DOI] [PubMed] [Google Scholar]
- Minciacchi V. R.; Freeman M. R.; Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015, 40, 41–51. 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M. Intercellular communication by exosomes in placenta: a possible role in cell fusion?. Placenta 2014, 35 (5), 297–302. 10.1016/j.placenta.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Yellon D. M.; Davidson S. M. Exosomes: nanoparticles involved in cardioprotection?. Circ. Res. 2014, 114 (2), 325–32. 10.1161/CIRCRESAHA.113.300636. [DOI] [PubMed] [Google Scholar]
- Henne W. M.; Buchkovich N. J.; Emr S. D. The ESCRT pathway. Dev Cell 2011, 21 (1), 77–91. 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Hurley J. H. ESCRTs are everywhere. EMBO J. 2015, 34 (19), 2398–407. 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya-Beltri C.; Baixauli F.; Gutierrez-Vazquez C.; Sanchez-Madrid F.; Mittelbrunn M. Sorting it out: regulation of exosome loading. Semin Cancer Biol. 2014, 28, 3–13. 10.1016/j.semcancer.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airola M. V.; Hannun Y. A. Sphingolipid metabolism and neutral sphingomyelinases. Handb Exp Pharmacol 2013, 215 (215), 57–76. 10.1007/978-3-7091-1368-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro B. M.; Prieto M.; Silva L. C. Ceramide: a simple sphingolipid with unique biophysical properties. Prog. Lipid Res. 2014, 54, 53–67. 10.1016/j.plipres.2014.01.004. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Liu Y.; Liu H.; Tang W. H. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. 10.1186/s13578-019-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batrakova E. V.; Kim M. S. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J. Controlled Release 2015, 219, 396–405. 10.1016/j.jconrel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A.; Costa-Silva B.; Shen T. L.; Rodrigues G.; Hashimoto A.; Tesic Mark M.; Molina H.; Kohsaka S.; Di Giannatale A.; Ceder S.; Singh S.; Williams C.; Soplop N.; Uryu K.; Pharmer L.; King T.; Bojmar L.; Davies A. E.; Ararso Y.; Zhang T.; Zhang H.; Hernandez J.; Weiss J. M.; Dumont-Cole V. D.; Kramer K.; Wexler L. H.; Narendran A.; Schwartz G. K.; Healey J. H.; Sandstrom P.; Labori K. J.; Kure E. H.; Grandgenett P. M.; Hollingsworth M. A.; de Sousa M.; Kaur S.; Jain M.; Mallya K.; Batra S. K.; Jarnagin W. R.; Brady M. S.; Fodstad O.; Muller V.; Pantel K.; Minn A. J.; Bissell M. J.; Garcia B. A.; Kang Y.; Rajasekhar V. K.; Ghajar C. M.; Matei I.; Peinado H.; Bromberg J.; Lyden D. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527 (7578), 329–335. 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulagnier K.; Vincent-Schneider H.; Hamdi S.; Subra C.; Lankar D.; Record M. Characterization of exosome subpopulations from RBL-2H3 cells using fluorescent lipids. Blood Cells Mol. Dis 2005, 35 (2), 116–21. 10.1016/j.bcmd.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Subra C.; Laulagnier K.; Perret B.; Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007, 89 (2), 205–12. 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Dou Y.; Cha D. J.; Franklin J. L.; Higginbotham J. N.; Jeppesen D. K.; Weaver A. M.; Prasad N.; Levy S.; Coffey R. J.; Patton J. G.; Zhang B. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci. Rep 2016, 6, 37982. 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowits G.; Gercel-Taylor C.; Day J. M.; Taylor D. D.; Kloecker G. H. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 2009, 10 (1), 42–6. 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- Yuan T.; Huang X.; Woodcock M.; Du M.; Dittmar R.; Wang Y.; Tsai S.; Kohli M.; Boardman L.; Patel T.; Wang L. Plasma extracellular RNA profiles in healthy and cancer patients. Sci. Rep 2016, 6, 19413. 10.1038/srep19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abels E. R.; Breakefield X. O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol. Neurobiol 2016, 36 (3), 301–12. 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gezer U.; Özgür E.; Cetinkaya M.; Isin M.; Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol. Int. 2014, 38 (9), 1076–1079. 10.1002/cbin.10301. [DOI] [PubMed] [Google Scholar]
- Jiang Y.; Wang F.; Wang K.; Zhong Y.; Wei X.; Wang Q.; Zhang H. Engineered Exosomes: A Promising Drug Delivery Strategy for Brain Diseases. Curr. Med. Chem. 2022, 29 (17), 3111–3124. 10.2174/0929867328666210902142015. [DOI] [PubMed] [Google Scholar]
- Goh C. Y.; Wyse C.; Ho M.; O’Beirne E.; Howard J.; Lindsay S.; Kelly P.; Higgins M.; McCann A. Exosomes in triple negative breast cancer: Garbage disposals or Trojan horses?. Cancer Lett. 2020, 473, 90–97. 10.1016/j.canlet.2019.12.046. [DOI] [PubMed] [Google Scholar]
- Zhou X.; Li T.; Chen Y.; Zhang N.; Wang P.; Liang Y.; Long M.; Liu H.; Mao J.; Liu Q.; Sun X.; Chen H. Mesenchymal stem cellderived extracellular vesicles promote the in vitro proliferation and migration of breast cancer cells through the activation of the ERK pathway. Int. J. Oncol. 2019, 54 (5), 1843–1852. 10.3892/ijo.2019.4747. [DOI] [PubMed] [Google Scholar]
- Wang S.; Li F.; Ye T.; Wang J.; Lyu C.; Qing S.; Ding Z.; Gao X.; Jia R.; Yu D.; Ren J.; Wei W.; Ma G. Macrophage-tumor chimeric exosomes accumulate in lymph node and tumor to activate the immune response and the tumor microenvironment. Sci. Transl Med. 2021, 13 (615), eabb6981 10.1126/scitranslmed.abb6981. [DOI] [PubMed] [Google Scholar]
- Lin S.; Yu Z.; Chen D.; Wang Z.; Miao J.; Li Q.; Zhang D.; Song J.; Cui D. Progress in Microfluidics-Based Exosome Separation and Detection Technologies for Diagnostic Applications. Small 2020, 16 (9), 1903916 10.1002/smll.201903916. [DOI] [PubMed] [Google Scholar]
- Chen L.; Wang L.; Zhu L.; Xu Z.; Liu Y.; Li Z.; Zhou J.; Luo F. Exosomes as Drug Carriers in Anti-Cancer Therapy. Front Cell Dev Biol. 2022, 10, 728616. 10.3389/fcell.2022.728616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L.; Yang X.; Gao Z.; Effah C. Y.; Zhang X.; Wu Y.; Qu L. A Holistic Review of the State-of-the-Art Microfluidics for Exosome Separation: An Overview of the Current Status, Existing Obstacles, and Future Outlook. Small 2021, 17 (29), 2007174 10.1002/smll.202007174. [DOI] [PubMed] [Google Scholar]
- Mohammadi M.; Zargartalebi H.; Salahandish R.; Aburashed R.; Wey Yong K.; Sanati-Nezhad A. Emerging technologies and commercial products in exosome-based cancer diagnosis and prognosis. Biosens Bioelectron 2021, 183, 113176. 10.1016/j.bios.2021.113176. [DOI] [PubMed] [Google Scholar]
- Narayanan E. Exosomes as drug delivery vehicles for cancer treatment. Current Nanoscience 2020, 16 (1), 15–26. 10.2174/1573413715666190219112422. [DOI] [Google Scholar]
- Wang J.; Ma P.; Kim D. H.; Liu B. F.; Demirci U. Towards Microfluidic-Based Exosome Isolation and Detection for Tumor Therapy. Nano Today 2021, 37, 101066. 10.1016/j.nantod.2020.101066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y.; Cheng G.; Liu X.; Hao S.-J.; Nisic M.; Zhu C.-D.; Xia Y.-Q.; Li W.-Q.; Wang Z.-G.; Zhang W.-L.; Rice S. J.; Sebastian A.; Albert I.; Belani C. P.; Zheng S.-Y. Rapid magnetic isolation of extracellular vesicles via lipid-based nanoprobes. Nat. Biomed Eng. 2017, 1, 0058. 10.1038/s41551-017-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y.; Maurer M.; He H. Z.; et al. Enrichment of extracellular vesicles with lipid nanoprobe functionalized nanostructured silica. Lab Chip 2019, 19 (14), 2346–2355. 10.1039/C8LC01359D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolatmoradi A.; Mirtaheri E.; El-Zahab B. Thermo-acoustofluidic separation of vesicles based on cholesterol content. Lab Chip 2017, 17 (7), 1332–1339. 10.1039/C7LC00161D. [DOI] [PubMed] [Google Scholar]
- Wu M.; Ozcelik A.; Rufo J.; Wang Z.; Fang R.; Jun Huang T. Acoustofluidic separation of cells and particles. Microsyst . Nanoeng. 2019, 5, 32. 10.1038/s41378-019-0064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Ji C.; Zhang H.; Shi H.; Mao F.; Qian H.; Xu W.; Wang D.; Pan J.; Fang X.; Santos H. A.; Zhang X. Engineered neutrophil-derived exosome-like vesicles for targeted cancer therapy. Sci. Adv. 2022, 8 (2), eabj8207 10.1126/sciadv.abj8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee I.; Syn N.; Sethi G.; Goh B. C.; Wang L. Role of tumor-derived exosomes in cancer metastasis. Biochim Biophys Acta Rev. Cancer 2019, 1871 (1), 12–19. 10.1016/j.bbcan.2018.10.004. [DOI] [PubMed] [Google Scholar]
- Jafari A.; Babajani A.; Abdollahpour-Alitappeh M.; Ahmadi N.; Rezaei-Tavirani M. Exosomes and cancer: from molecular mechanisms to clinical applications. Med. Oncol. 2021, 38, 45. 10.1007/s12032-021-01491-0. [DOI] [PubMed] [Google Scholar]
- Huang Y. K.; Wang M.; Sun Y.; Di Costanzo N.; Mitchell C.; Achuthan A.; Hamilton J. A.; Busuttil R. A.; Boussioutas A. Macrophage spatial heterogeneity in gastric cancer defined by multiplex immunohistochemistry. Nat. Commun. 2019, 10, 3928. 10.1038/s41467-019-11788-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X.; Wang S.; Sun M.; Zhang C.; Wei C.; Yang C.; Dou R.; Liu Q.; Xiong B. Correction to: miR-195–5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J. Hematol. Oncol. 2019, 12, 122. 10.1186/s13045-019-0810-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C.; Zhang C.; Wang H.; Zhao L. Exosome-mediated communication between tumor cells and tumor-associated macrophages: implications for tumor microenvironment. Oncoimmunology 2021, 10 (1), 1887552. 10.1080/2162402X.2021.1887552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.; Deep G. Hypoxia in tumor microenvironment regulates exosome biogenesis: Molecular mechanisms and translational opportunities. Cancer Lett. 2020, 479, 23–30. 10.1016/j.canlet.2020.03.017. [DOI] [PubMed] [Google Scholar]
- Park J. E.; Dutta B.; Tse S. W.; Gupta N.; Tan C. F.; Low J. K.; Yeoh K. W.; Kon O. L.; Tam J. P.; Sze S. K. Hypoxia-induced tumor exosomes promote M2-like macrophage polarization of infiltrating myeloid cells and microRNA-mediated metabolic shift. Oncogene 2019, 38 (26), 5158–5173. 10.1038/s41388-019-0782-x. [DOI] [PubMed] [Google Scholar]
- Luan Y.; Li X.; Luan Y.; Zhao R.; Li Y.; Liu L.; Hao Y.; Oleg Vladimir B.; Jia L. Circulating lncRNA UCA1 Promotes Malignancy of Colorectal Cancer via the miR-143/MYO6 Axis. Mol. Ther Nucleic Acids 2020, 19, 790–803. 10.1016/j.omtn.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Qin X.; Bian W.; Li Y.; Shan B.; Yao Z.; Li S. Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA-124/STAT3 axis. J. Exp. Clin. Cancer Res. 2019, 38, 477. 10.1186/s13046-019-1473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltesz B.; Buglyo G.; Nemeth N.; Szilagyi M.; Pos O.; Szemes T.; Balogh I.; Nagy B. The Role of Exosomes in Cancer Progression. Int. J. Mol. Sci. 2022, 23 (1), 8. 10.3390/ijms23010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi A.; Mehic J.; Creskey M.; Gobin J.; Gao J.; Rigg E.; Muradia G.; Luebbert C. C.; Westwood C.; Stalker A.; Allan D. S.; Johnston M. J. W.; Cyr T.; Rosu-Myles M.; Lavoie J. R. A comprehensive proteomics profiling identifies NRP1 as a novel identity marker of human bone marrow mesenchymal stromal cell-derived small extracellular vesicles. Stem Cell Res. Ther. 2019, 10, 401. 10.1186/s13287-019-1516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Denis-Bissonnette F.; Khoury R.; Mediratta K.; El-Sahli S.; Wang L.; Lavoie J. R. Applications of Extracellular Vesicles in Triple-Negative Breast Cancer. Cancers (Basel) 2022, 14 (2), 451. 10.3390/cancers14020451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Ricardo J.; Leal-Orta E.; Martinez-Baeza E.; Ortiz-Mendoza C.; Breton-Mora F.; Herrera-Torres A.; Elizalde-Acosta I.; Cortes-Reynosa P.; Thompson-Bonilla R.; Perez Salazar E. Circulating extracellular vesicles from patients with breast cancer enhance migration and invasion via a Srcdependent pathway in MDAMB231 breast cancer cells. Mol. Med. Rep 2020, 22 (3), 1932–1948. 10.3892/mmr.2020.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe D. C.; Walker N. D.; Rustandy F. D.; Wallace J.; Lee J.; Stott S. L.; Rosner M. R. Tumor Extracellular Vesicles Regulate Macrophage-Driven Metastasis through CCL5. Cancers 2021, 13 (14), 3459. 10.3390/cancers13143459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. X.; Gires O. Tumor-derived extracellular vesicles in breast cancer: From bench to bedside. Cancer Lett. 2019, 460, 54–64. 10.1016/j.canlet.2019.06.012. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M.; Iinuma H.; Umemoto Y.; Yanagisawa T.; Matsumoto A.; Jinno H. Exosome-encapsulated microRNA-223–3p as a minimally invasive biomarker for the early detection of invasive breast cancer. Oncol Lett. 2018, 15 (6), 9584–9592. 10.3892/ol.2018.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.; Chen H.; Xu C.; et al. Exosomal miR-106b serves as a novel marker for lung cancer and promotes cancer metastasis via targeting PTEN. Life Sci. 2020, 244, 117297. 10.1016/j.lfs.2020.117297. [DOI] [PubMed] [Google Scholar]
- Tian Y.; Ma L.; Gong M.; et al. Protein Profiling and Sizing of Extracellular Vesicles from Colorectal Cancer Patients via Flow Cytometry. ACS Nano 2018, 12 (1), 671–680. 10.1021/acsnano.7b07782. [DOI] [PubMed] [Google Scholar]
- Rodriguez M.; Bajo-Santos C.; Hessvik N. P.; Lorenz S.; Fromm B.; Berge V.; Sandvig K.; Line̅ A.; Llorente A.; et al. Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol. Cancer 2017, 16, 156. 10.1186/s12943-017-0726-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.-Y.; Huang Z.-L.; Huang J.; Xu B.; Huang X.-Y.; Xu Y.-H.; Zhou J.; Tang Z.-Y.; et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 2020, 39, 20. 10.1186/s13046-020-1529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkhoury K.; Kocak P.; Kang A.; Arab-Tehrany E.; Ellis Ward J.; Shin S. R. Engineering Smart Targeting Nanovesicles and Their Combination with Hydrogels for Controlled Drug Delivery. Pharmaceutics 2020, 12 (9), 849. 10.3390/pharmaceutics12090849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butreddy A.; Kommineni N.; Dudhipala N. Exosomes as Naturally Occurring Vehicles for Delivery of Biopharmaceuticals: Insights from Drug Delivery to Clinical Perspectives. Nanomaterials 2021, 11 (6), 1481. 10.3390/nano11061481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Ríos A. J.; Molina-Crespo Á.; Bouzo B. L.; López-López R.; Moreno-Bueno G.; de la Fuente M. Exosome-mimetic nanoplatforms for targeted cancer drug delivery. J. Nanobiotechnology 2019, 17, 85. 10.1186/s12951-019-0517-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vigani B.; Rossi S.; Sandri G.; Bonferoni M. C.; Caramella C. M.; Ferrari F. Recent Advances in the Development of In Situ Gelling Drug Delivery Systems for Non-Parenteral Administration Routes. Pharmaceutics 2020, 12 (9), 859. 10.3390/pharmaceutics12090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibria G.; Ramos E. K.; Wan Y.; Gius D. R.; Liu H. Exosomes as a Drug Delivery System in Cancer Therapy: Potential and Challenges. Mol. Pharmaceutics 2018, 15 (9), 3625–3633. 10.1021/acs.molpharmaceut.8b00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J.; Zaro J.; Shen Y. Advances in Exosome-Based Drug Delivery and Tumor Targeting: From Tissue Distribution to Intracellular Fate. Int. J. Nanomedicine 2020, 15, 9355–9371. 10.2147/IJN.S281890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Millan C.; Calvo Díaz C.; Lanao J. M.; Colino C. I. Advances in exosomes-based drug delivery systems. Macromol. Biosci. 2021, 21 (1), 2000269. 10.1002/mabi.202000269. [DOI] [PubMed] [Google Scholar]
- Kučuk N.; Primožič M.; Knez Ž.; Leitgeb M. Exosomes Engineering and Their Roles as Therapy Delivery Tools, Therapeutic Targets, and Biomarkers. Int. J. Mol. Sci. 2021, 22 (17), 9543. 10.3390/ijms22179543. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li Y.; Zhang Y.; Li Z.; Zhou K.; Feng N. Exosomes as Carriers for Antitumor Therapy. ACS Biomater Sci. Eng. 2019, 5 (10), 4870–4881. 10.1021/acsbiomaterials.9b00417. [DOI] [PubMed] [Google Scholar]
- Vader P.; Mol E. A.; Pasterkamp G.; Schiffelers R. M. Extracellular vesicles for drug delivery. Adv. Drug Deliv. Rev. 2016, 106, 148–156. 10.1016/j.addr.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Akuma P.; Okagu O. D.; Udenigwe C. C. Naturally occurring exosome vesicles as potential delivery vehicle for bioactive compounds. Front. Sustain. Food Syst. 2019, 3, 23. 10.3389/fsufs.2019.00023. [DOI] [Google Scholar]
- Butreddy A.; Kommineni N.; Dudhipala N. Exosomes as Naturally Occurring Vehicles for Delivery of Biopharmaceuticals: Insights from Drug Delivery to Clinical Perspectives. Nanomaterials 2021, 11 (6), 1481. 10.3390/nano11061481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann G.; Serio A.; Mazo M.; Nair R.; Stevens M. M. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J. Controlled Release 2015, 205, 35–44. 10.1016/j.jconrel.2014.11.029. [DOI] [PubMed] [Google Scholar]
- Kim M. S.; Haney M. J.; Zhao Y.; Mahajan V.; Deygen I.; Klyachko N. L.; Inskoe E.; Piroyan A.; Sokolsky M.; Okolie O.; Hingtgen S. D.; Kabanov A. V.; Batrakova E. V. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine 2016, 12 (3), 655–664. 10.1016/j.nano.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y. T.; Umezaki K.; Sawada S.; Mukai S. A.; Sasaki Y.; Harada N.; Shiku H.; Akiyoshi K. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep 2016, 6, 21933. 10.1038/srep21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney M. J.; Klyachko N. L.; Zhao Y.; Gupta R.; Plotnikova E. G.; He Z.; Patel T.; Piroyan A.; Sokolsky M.; Kabanov A. V.; Batrakova E. V. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Controlled Release 2015, 207, 18–30. 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Y.; Li X.; Luan Y.; Zhao R.; Li Y.; Liu L.; Hao Y.; Oleg Vladimir B.; Jia L. Circulating lncRNA UCA1 Promotes Malignancy of Colorectal Cancer via the miR-143/MYO6 Axis. Mol. Ther Nucleic Acids 2020, 19, 790–803. 10.1016/j.omtn.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.; Jang H.; Cho H.; Choi J.; Hwang K. Y.; Choi Y.; Kim S. H.; Yang Y. Recent Advances in Exosome-Based Drug Delivery for Cancer Therapy. Cancers 2021, 13 (17), 4435. 10.3390/cancers13174435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung S.; Perocheau D.; Touramanidou L.; Baruteau J. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun. Signal 2021, 19, 47. 10.1186/s12964-021-00730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camussi G.; Deregibus M. C.; Bruno S.; Grange C.; Fonsato V.; Tetta C. Exosome/microvesicle-mediated epigenetic reprogramming of cells. Am. J. Cancer Res. 2011, 1 (1), 98–110. [PMC free article] [PubMed] [Google Scholar]
- Bruno S.; Grange C.; Deregibus M. C.; Calogero R. A.; Saviozzi S.; Collino F.; Morando L.; Busca A.; Falda M.; Bussolati B.; Tetta C.; Camussi G. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J. Am. Soc. Nephrol 2009, 20 (5), 1053–67. 10.1681/ASN.2008070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsharkasy O. M.; Nordin J. Z.; Hagey D. W.; de Jong O. G.; Schiffelers R. M.; Andaloussi S. E.; Vader P. Extracellular vesicles as drug delivery systems: Why and how?. Adv. Drug Deliv Rev. 2020, 159, 332–343. 10.1016/j.addr.2020.04.004. [DOI] [PubMed] [Google Scholar]
- Armstrong J. P.; Holme M. N.; Stevens M. M. Re-Engineering Extracellular Vesicles as Smart Nanoscale Therapeutics. ACS Nano 2017, 11 (1), 69–83. 10.1021/acsnano.6b07607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara P.; Chan A. B.; Cruz L. J.; Quest A. F. G.; Kogan M. J. Exploiting the Natural Properties of Extracellular Vesicles in Targeted Delivery towards Specific Cells and Tissues. Pharmaceutics 2020, 12 (11), 1022. 10.3390/pharmaceutics12111022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.; Zhuang X.; Xiang X.; Liu Y.; Zhang S.; Liu C.; Barnes S.; Grizzle W.; Miller D.; Zhang H. G. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol. Ther 2010, 18 (9), 1606–14. 10.1038/mt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks W. A.; Sharma P.; Bullock K. M.; Hansen K. M.; Ludwig N.; Whiteside T. L. Transport of Extracellular Vesicles across the Blood-Brain Barrier: Brain Pharmacokinetics and Effects of Inflammation. Int. J. Mol. Sci. 2020, 21 (12), 4407. 10.3390/ijms21124407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.; Zheng T.; Zhang B. Exosomes in Parkinson’s Disease. Neurosci Bull. 2017, 33 (3), 331–338. 10.1007/s12264-016-0092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.; Yim H.; Park C.; Ahn S. H.; Ahn Y.; Lee A.; Yang H.; Choi C. Targeted Delivery of Exosomes Armed with Anti-Cancer Therapeutics. Membranes 2022, 12 (1), 85. 10.3390/membranes12010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Li S.; Li L.; Li M.; Guo C.; Yao J.; Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 2015, 13 (1), 17–24. 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z.; Shi K.; Yang S.; Liu J.; Zhou Q.; Wang G.; Song J.; Li Z.; Zhang Z.; Yuan W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol. Cancer 2018, 17, 147. 10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J.; Tkach M.; Thery C. Biogenesis and secretion of exosomes. Curr. Opin Cell Biol. 2014, 29, 116–25. 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- Tai Y. L.; Chen K. C.; Hsieh J. T.; Shen T. L. Exosomes in cancer development and clinical applications. Cancer Sci. 2018, 109 (8), 2364–2374. 10.1111/cas.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A.; Thakur B. K.; Weiss J. M.; Kim H. S.; Peinado H.; Lyden D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30 (6), 836–848. 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quah B. J.; O’Neill H. C. The immunogenicity of dendritic cell-derived exosomes. Blood Cells Mol. Dis 2005, 35 (2), 94–110. 10.1016/j.bcmd.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Zheng M.; Huang M.; Ma X.; Chen H.; Gao X. Harnessing Exosomes for the Development of Brain Drug Delivery Systems. Bioconjug Chem. 2019, 30 (4), 994–1005. 10.1021/acs.bioconjchem.9b00085. [DOI] [PubMed] [Google Scholar]
- Jiang B.; Kauffman A. E.; Li L.; McFee W.; Cai B.; Weinstein J.; Lead J. R.; Chatterjee S.; Scott G. I.; Xiao S. Health impacts of environmental contamination of micro- and nanoplastics: a review. Environ. Health Prev. Med. 2020, 25, 29. 10.1186/s12199-020-00870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y.; Li S.; Song J.; Ji T.; Zhu M.; Anderson G. J.; Wei J.; Nie G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014, 35 (7), 2383–90. 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- Bai J.; Duan J.; Liu R.; Du Y.; Luo Q.; Cui Y.; Su Z.; Xu J.; Xie Y.; Lu W. Engineered targeting tLyp-1 exosomes as gene therapy vectors for efficient delivery of siRNA into lung cancer cells. Asian J. Pharm. Sci. 2020, 15 (4), 461–471. 10.1016/j.ajps.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellavia D.; Raimondo S.; Calabrese G.; Forte S.; Cristaldi M.; Patinella A.; Memeo L.; Manno M.; Raccosta S.; Diana P.; Cirrincione G.; Giavaresi G.; Monteleone F.; Fontana S.; De Leo G.; Alessandro R. Interleukin 3- receptor targeted exosomes inhibit in vitro and in vivo Chronic Myelogenous Leukemia cell growth. Theranostics 2017, 7 (5), 1333–1345. 10.7150/thno.17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G.; Zhu Y.; Ali D. J.; Tian T.; Xu H.; Si K.; Sun B.; Chen B.; Xiao Z. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J. Nanobiotechnol. 2020, 18, 10. 10.1186/s12951-019-0563-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoni S. K.; Moghadam M. F.; Moazzeni S. M.; Gomari H.; Salimi F. Engineered Exosomes for Targeted Transfer of siRNA to HER2 Positive Breast Cancer Cells. Appl. Biochem. Biotechnol. 2019, 187 (1), 352–364. 10.1007/s12010-018-2813-4. [DOI] [PubMed] [Google Scholar]
- Ohno S.; Takanashi M.; Sudo K.; Ueda S.; Ishikawa A.; Matsuyama N.; Fujita K.; Mizutani T.; Ohgi T.; Ochiya T.; Gotoh N.; Kuroda M. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther 2013, 21 (1), 185–91. 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q.; Shi X.; Han M.; Smbatyan G.; Lenz H. J.; Zhang Y. Reprogramming Exosomes as Nanoscale Controllers of Cellular Immunity. J. Am. Chem. Soc. 2018, 140 (48), 16413–16417. 10.1021/jacs.8b10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G.; Kan S.; Zhu Y.; Feng S.; Feng W.; Gao S. Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int. J. Nanomedicine 2018, 13, 585–599. 10.2147/IJN.S154458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanuma T.; Yamamoto T.; Kobiyama K.; Moriishi E.; Masuta Y.; Kusakabe T.; Ozasa K.; Kuroda E.; Jounai N.; Ishii K. J. CD63-Mediated Antigen Delivery into Extracellular Vesicles via DNA Vaccination Results in Robust CD8(+) T Cell Responses. J. Immunol 2017, 198 (12), 4707–4715. 10.4049/jimmunol.1600731. [DOI] [PubMed] [Google Scholar]
- Rountree R. B.; Mandl S. J.; Nachtwey J. M.; Dalpozzo K.; Do L.; Lombardo J. R.; Schoonmaker P. L.; Brinkmann K.; Dirmeier U.; Laus R.; Delcayre A. Exosome targeting of tumor antigens expressed by cancer vaccines can improve antigen immunogenicity and therapeutic efficacy. Cancer Res. 2011, 71 (15), 5235–44. 10.1158/0008-5472.CAN-10-4076. [DOI] [PubMed] [Google Scholar]
- Wang M.; Altinoglu S.; Takeda Y. S.; Xu Q. Integrating Protein Engineering and Bioorthogonal Click Conjugation for Extracellular Vesicle Modulation and Intracellular Delivery. PLoS One 2015, 10 (11), e0141860 10.1371/journal.pone.0141860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G.; Han Y.; An Y.; Ding Y.; He C.; Wang X.; Tang Q. NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomaterials 2018, 178, 302–316. 10.1016/j.biomaterials.2018.06.029. [DOI] [PubMed] [Google Scholar]
- Kim M. S.; Haney M. J.; Zhao Y.; Yuan D.; Deygen I.; Klyachko N. L.; Kabanov A. V.; Batrakova E. V. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: in vitro and in vivo evaluations. Nanomedicine 2018, 14 (1), 195–204. 10.1016/j.nano.2017.09.011. [DOI] [PubMed] [Google Scholar]
- Cao Y.; Wu T.; Zhang K.; Meng X.; Dai W.; Wang D.; Dong H.; Zhang X. Engineered Exosome-Mediated Near-Infrared-II Region V2C Quantum Dot Delivery for Nucleus-Target Low-Temperature Photothermal Therapy. ACS Nano 2019, 13 (2), 1499–1510. 10.1021/acsnano.8b07224. [DOI] [PubMed] [Google Scholar]
- Pi F.; Binzel D. W.; Lee T. J.; Li Z.; Sun M.; Rychahou P.; Li H.; Haque F.; Wang S.; Croce C. M.; Guo B.; Evers B. M.; Guo P. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat. Nanotechnol 2018, 13 (1), 82–89. 10.1038/s41565-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J.; Shi M.; Liu X.; Jin C.; Xing X.; Qiu L.; Tan W. Aptamer-Functionalized Exosomes: Elucidating the Cellular Uptake Mechanism and the Potential for Cancer-Targeted Chemotherapy. Anal. Chem. 2019, 91 (3), 2425–2430. 10.1021/acs.analchem.8b05204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y.; Wang L.; Zhu C.; Zheng Q.; Wang G.; Tong J.; Fang Y.; Xia Y.; Cheng G.; He X.; Zheng S. Y. Aptamer-Conjugated Extracellular Nanovesicles for Targeted Drug Delivery. Cancer Res. 2018, 78 (3), 798–808. 10.1158/0008-5472.CAN-17-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z.; Zhang T.; He W.; Jin H.; Liu C.; Yang Z.; Ren J. Methotrexate-Loaded Extracellular Vesicles Functionalized with Therapeutic and Targeted Peptides for the Treatment of Glioblastoma Multiforme. ACS Appl. Mater. Interfaces 2018, 10 (15), 12341–12350. 10.1021/acsami.7b18135. [DOI] [PubMed] [Google Scholar]
- Cheng H.; Fan J. H.; Zhao L. P.; Fan G. L.; Zheng R. R.; Qiu X. Z.; Yu X. Y.; Li S. Y.; Zhang X. Z. Chimeric peptide engineered exosomes for dual-stage light guided plasma membrane and nucleus targeted photodynamic therapy. Biomaterials 2019, 211, 14–24. 10.1016/j.biomaterials.2019.05.004. [DOI] [PubMed] [Google Scholar]
- Koh E.; Lee E. J.; Nam G. H.; Hong Y.; Cho E.; Yang Y.; Kim I. S. Exosome-SIRPalpha, a CD47 blockade increases cancer cell phagocytosis. Biomaterials 2017, 121, 121–129. 10.1016/j.biomaterials.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Wang J.; Li W.; Lu Z.; Zhang L.; Hu Y.; Li Q.; Du W.; Feng X.; Jia H.; Liu B. F. The use of RGD-engineered exosomes for enhanced targeting ability and synergistic therapy toward angiogenesis. Nanoscale 2017, 9 (40), 15598–15605. 10.1039/C7NR04425A. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Shi J.; Xie J.; Wang Y.; Sun J.; Liu T.; Zhao Y.; Zhao X.; Wang X.; Ma Y.; Malkoc V.; Chiang C.; Deng W.; Chen Y.; Fu Y.; Kwak K. J.; Fan Y.; Kang C.; Yin C.; Rhee J.; Bertani P.; Otero J.; Lu W.; Yun K.; Lee A. S.; Jiang W.; Teng L.; Kim B. Y. S.; Lee L. J. Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nat. Biomed Eng. 2020, 4 (1), 69–83. 10.1038/s41551-019-0485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S.; Li T.; Wen X.; Wu S. Y.; Xiong C.; Zhao J.; Lincha V. R.; Chow D. S.; Liu Y.; Sood A. K.; Li C. Copper-64 Labeled PEGylated Exosomes for In Vivo Positron Emission Tomography and Enhanced Tumor Retention. Bioconjug Chem. 2019, 30 (10), 2675–2683. 10.1021/acs.bioconjchem.9b00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Wu Y.; Ding F.; Yang J.; Li J.; Gao X.; Zhang C.; Feng J. Engineering macrophage-derived exosomes for targeted chemotherapy of triple-negative breast cancer. Nanoscale 2020, 12 (19), 10854–10862. 10.1039/D0NR00523A. [DOI] [PubMed] [Google Scholar]
- Ortega A.; Martinez-Arroyo O.; Forner M. J.; Cortes R. Exosomes as Drug Delivery Systems: Endogenous Nanovehicles for Treatment of Systemic Lupus Erythematosus. Pharmaceutics 2021, 13 (1), 3. 10.3390/pharmaceutics13010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L.; Gu Y.; Du Y.; Liu J. Exosomes: Diagnostic Biomarkers and Therapeutic Delivery Vehicles for Cancer. Mol. Pharmaceutics 2019, 16 (8), 3333–3349. 10.1021/acs.molpharmaceut.9b00409. [DOI] [PubMed] [Google Scholar]
- von Minckwitz G.; Huang C.-S.; Mano M. S.; Loibl S.; Mamounas E. P.; Untch M.; Wolmark N.; Rastogi P.; Schneeweiss A.; Redondo A.; Fischer H. H.; Jacot W.; Conlin A. K.; Arce-Salinas C.; Wapnir I. L.; Jackisch C.; DiGiovanna M. P.; Fasching P. A.; Crown J. P.; Wulfing P.; Shao Z.; Rota Caremoli E.; Wu H.; Lam L. H.; Tesarowski D.; Smitt M.; Douthwaite H.; Singel S. M.; Geyer C. E. Trastuzumab Emtansine for Residual Invasive HER2-Positive Breast Cancer. N. Engl. J. Med. 2019, 380 (7), 617–628. 10.1056/NEJMoa1814017. [DOI] [PubMed] [Google Scholar]
- Schmid P.; Adams S.; Rugo H. S.; Schneeweiss A.; Barrios C. H.; Iwata H.; Dieras V.; Hegg R.; Im S. A.; Shaw Wright G.; Henschel V.; Molinero L.; Chui S. Y.; Funke R.; Husain A.; Winer E. P.; Loi S.; Emens L. A. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379 (22), 2108–2121. 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- Im S. A.; Lu Y. S.; Bardia A.; Harbeck N.; Colleoni M.; Franke F.; Chow L.; Sohn J.; Lee K. S.; Campos-Gomez S.; Villanueva-Vazquez R.; Jung K. H.; Chakravartty A.; Hughes G.; Gounaris I.; Rodriguez-Lorenc K.; Taran T.; Hurvitz S.; Tripathy D. Overall Survival with Ribociclib plus Endocrine Therapy in Breast Cancer. N Engl J. Med. 2019, 381 (4), 307–316. 10.1056/NEJMoa1903765. [DOI] [PubMed] [Google Scholar]
- Horn L.; Mansfield A. S.; Szczesna A.; Havel L.; Krzakowski M.; Hochmair M. J.; Huemer F.; Losonczy G.; Johnson M. L.; Nishio M.; Reck M.; Mok T.; Lam S.; Shames D. S.; Liu J.; Ding B.; Lopez-Chavez A.; Kabbinavar F.; Lin W.; Sandler A.; Liu S. V. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 379 (23), 2220–2229. 10.1056/NEJMoa1809064. [DOI] [PubMed] [Google Scholar]
- Van Cutsem E.; Huijberts S.; Grothey A.; Yaeger R.; Cuyle P. J.; Elez E.; Fakih M.; Montagut C.; Peeters M.; Yoshino T.; Wasan H.; Desai J.; Ciardiello F.; Gollerkeri A.; Christy-Bittel J.; Maharry K.; Sandor V.; Schellens J. H. M.; Kopetz S.; Tabernero J. Binimetinib, Encorafenib, and Cetuximab Triplet Therapy for Patients With BRAF V600E-Mutant Metastatic Colorectal Cancer: Safety Lead-In Results From the Phase III BEACON Colorectal Cancer Study. J. Clin Oncol 2019, 37 (17), 1460–1469. 10.1200/JCO.18.02459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I. D.; Martin A. J.; Stockler M. R.; Begbie S.; Chi K. N.; Chowdhury S.; Coskinas X.; Frydenberg M.; Hague W. E.; Horvath L. G.; Joshua A. M.; Lawrence N. J.; Marx G.; McCaffrey J.; McDermott R.; McJannett M.; North S. A.; Parnis F.; Parulekar W.; Pook D. W.; Reaume M. N.; Sandhu S. K.; Tan A.; Tan T. H.; Thomson A.; Tu E.; Vera-Badillo F.; Williams S. G.; Yip S.; Zhang A. Y.; Zielinski R. R.; Sweeney C. J. Enzalutamide with Standard First-Line Therapy in Metastatic Prostate Cancer. N. Engl. J. Med. 2019, 381 (2), 121–131. 10.1056/NEJMoa1903835. [DOI] [PubMed] [Google Scholar]
- Rini B. I.; Plimack E. R.; Stus V.; Gafanov R.; Hawkins R.; Nosov D.; Pouliot F.; Alekseev B.; Soulieres D.; Melichar B.; Vynnychenko I.; Kryzhanivska A.; Bondarenko I.; Azevedo S. J.; Borchiellini D.; Szczylik C.; Markus M.; McDermott R. S.; Bedke J.; Tartas S.; Chang Y. H.; Tamada S.; Shou Q.; Perini R. F.; Chen M.; Atkins M. B.; Powles T. Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2019, 380 (12), 1116–1127. 10.1056/NEJMoa1816714. [DOI] [PubMed] [Google Scholar]
- Motzer R. J.; Penkov K.; Haanen J.; Rini B.; Albiges L.; Campbell M. T.; Venugopal B.; Kollmannsberger C.; Negrier S.; Uemura M.; Lee J. L.; Vasiliev A.; Miller W. H. Jr.; Gurney H.; Schmidinger M.; Larkin J.; Atkins M. B.; Bedke J.; Alekseev B.; Wang J.; Mariani M.; Robbins P. B.; Chudnovsky A.; Fowst C.; Hariharan S.; Huang B.; di Pietro A.; Choueiri T. K. Avelumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J. Med. 2019, 380 (12), 1103–1115. 10.1056/NEJMoa1816047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrlinger U.; Tzaridis T.; Mack F.; Steinbach J. P.; Schlegel U.; Sabel M.; Hau P.; Kortmann R. D.; Krex D.; Grauer O.; Goldbrunner R.; Schnell O.; Bahr O.; Uhl M.; Seidel C.; Tabatabai G.; Kowalski T.; Ringel F.; Schmidt-Graf F.; Suchorska B.; Brehmer S.; Weyerbrock A.; Renovanz M.; Bullinger L.; Galldiks N.; Vajkoczy P.; Misch M.; Vatter H.; Stuplich M.; Schafer N.; Kebir S.; Weller J.; Schaub C.; Stummer W.; Tonn J. C.; Simon M.; Keil V. C.; Nelles M.; Urbach H.; Coenen M.; Wick W.; Weller M.; Fimmers R.; Schmid M.; Hattingen E.; Pietsch T.; Coch C.; Glas M. Neurooncology Working Group of the German Cancer, S., Lomustine-Temozolomide combination therapy versus standard Temozolomide therapy in patients with newly diagnosed glioblastoma with methylated MGMT promoter (CeTeG/NOA-09): a randomised, open-label, phase 3 trial. Lancet 2019, 393 (10172), 678–688. 10.1016/S0140-6736(18)31791-4. [DOI] [PubMed] [Google Scholar]
- Fangusaro J.; Onar-Thomas A.; Young Poussaint T.; Wu S.; Ligon A. H.; Lindeman N.; Banerjee A.; Packer R. J.; Kilburn L. B.; Goldman S.; Pollack I. F.; Qaddoumi I.; Jakacki R. I.; Fisher P. G.; Dhall G.; Baxter P.; Kreissman S. G.; Stewart C. F.; Jones D. T. W.; Pfister S. M.; Vezina G.; Stern J. S.; Panigrahy A.; Patay Z.; Tamrazi B.; Jones J. Y.; Haque S. S.; Enterline D. S.; Cha S.; Fisher M. J.; Doyle L. A.; Smith M.; Dunkel I. J.; Fouladi M. Selumetinib in paediatric patients with BRAF-aberrant or neurofibromatosis type 1-associated recurrent, refractory, or progressive low-grade glioma: a multicentre, phase 2 trial. Lancet Oncol 2019, 20 (7), 1011–1022. 10.1016/S1470-2045(19)30277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard J. P.; Trneny M.; Izutsu K.; Fowler N. H.; Hong X.; Zhu J.; Zhang H.; Offner F.; Scheliga A.; Nowakowski G. S.; Pinto A.; Re F.; Fogliatto L. M.; Scheinberg P.; Flinn I. W.; Moreira C.; Cabecadas J.; Liu D.; Kalambakas S.; Fustier P.; Wu C.; Gribben J. G. AUGMENT: A Phase III Study of Lenalidomide Plus Rituximab Versus Placebo Plus Rituximab in Relapsed or Refractory Indolent Lymphoma. J. Clin Oncol 2019, 37 (14), 1188–1199. 10.1200/JCO.19.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter S. S.; Dunsmore K. P.; Devidas M.; Wood B. L.; Esiashvili N.; Chen Z.; Eisenberg N.; Briegel N.; Hayashi R. J.; Gastier-Foster J. M.; Carroll A. J.; Heerema N. A.; Asselin B. L.; Gaynon P. S.; Borowitz M. J.; Loh M. L.; Rabin K. R.; Raetz E. A.; Zweidler-Mckay P. A.; Winick N. J.; Carroll W. L.; Hunger S. P. Improved Survival for Children and Young Adults With T-Lineage Acute Lymphoblastic Leukemia: Results From the Children’s Oncology Group AALL0434 Methotrexate Randomization. J. Clin Oncol 2018, 36 (29), 2926–2934. 10.1200/JCO.2018.77.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzenstein H. M.; Langham M. R.; Malogolowkin M. H.; Krailo M. D.; Towbin A. J.; McCarville M. B.; Finegold M. J.; Ranganathan S.; Dunn S.; McGahren E. D.; Tiao G. M.; O’Neill A. F.; Qayed M.; Furman W. L.; Xia C.; Rodriguez-Galindo C.; Meyers R. L. Minimal adjuvant chemotherapy for children with hepatoblastoma resected at diagnosis (AHEP0731): a Children’s Oncology Group, multicentre, phase 3 trial. Lancet Oncol 2019, 20 (5), 719–727. 10.1016/S1470-2045(18)30895-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long G. V.; Saw R. P. M.; Lo S.; Nieweg O. E.; Shannon K. F.; Gonzalez M.; Guminski A.; Lee J. H.; Lee H.; Ferguson P. M.; Rawson R. V.; Wilmott J. S.; Thompson J. F.; Kefford R. F.; Ch’ng S.; Stretch J. R.; Emmett L.; Kapoor R.; Rizos H.; Spillane A. J.; Scolyer R. A.; Menzies A. M. Neoadjuvant dabrafenib combined with trametinib for resectable, stage IIIB-C, BRAF(V600) mutation-positive melanoma (NeoCombi): a single-arm, open-label, single-centre, phase 2 trial. Lancet Oncol 2019, 20 (7), 961–971. 10.1016/S1470-2045(19)30331-6. [DOI] [PubMed] [Google Scholar]
- Rozeman E. A.; Menzies A. M.; van Akkooi A. C. J.; Adhikari C.; Bierman C.; van de Wiel B. A.; Scolyer R. A.; Krijgsman O.; Sikorska K.; Eriksson H.; Broeks A.; van Thienen J. V.; Guminski A. D.; Acosta A. T.; Ter Meulen S.; Koenen A. M.; Bosch L. J. W.; Shannon K.; Pronk L. M.; Gonzalez M.; Ch’ng S.; Grijpink-Ongering L. G.; Stretch J.; Heijmink S.; van Tinteren H.; Haanen J.; Nieweg O. E.; Klop W. M. C.; Zuur C. L.; Saw R. P. M.; van Houdt W. J.; Peeper D. S.; Spillane A. J.; Hansson J.; Schumacher T. N.; Long G. V.; Blank C. U. Identification of the optimal combination dosing schedule of neoadjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma (OpACIN-neo): a multicentre, phase 2, randomised, controlled trial. Lancet Oncol 2019, 20 (7), 948–960. 10.1016/S1470-2045(19)30151-2. [DOI] [PubMed] [Google Scholar]
- Dhar R.; Mallik S.; Devi A. Exosomal microRNAs (exoMIRs): micromolecules with macro impact in oral cancer. 3 Biotech 2022, 12 (7), 155. 10.1007/s13205-022-03217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. J.; Wu J. Y.; Wang J. M.; Hu X. B.; Cai J. X.; Xiang D. X. Gemcitabine loaded autologous exosomes for effective and safe chemotherapy of pancreatic cancer. Acta Biomater. 2020, 101, 519–530. 10.1016/j.actbio.2019.10.022. [DOI] [PubMed] [Google Scholar]
- Smyth T.; Kullberg M.; Malik N.; Smith-Jones P.; Graner M. W.; Anchordoquy T. J. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J. Controlled Release 2015, 199, 145–155. 10.1016/j.jconrel.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T.; Takahashi Y.; Nishikawa M.; Kato K.; Morishita M.; Yamashita T.; Matsumoto A.; Charoenviriyakul C.; Takakura Y. Macrophage-dependent clearance of systemically administered B16BL6-derived exosomes from the blood circulation in mice. J. Extracell. Vesicles 2015, 4 (1), 26238. 10.3402/jev.v4.26238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijmans S.A.A.; Fliervoet L.A.L.; van der Meel R.; Fens M.H.A.M.; Heijnen H.F.G.; van Bergen en Henegouwen P.M.P.; Vader P.; Schiffelers R.M. PEGylated and targeted extracellular vesicles display enhanced cell specificity and circulation time. J. Controlled Release 2016, 224, 77–85. 10.1016/j.jconrel.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Shivji G. G.; Dhar R.; Devi A. Role of exosomes and its emerging therapeutic applications in the pathophysiology of non-infectious diseases. Biomarkers 2022, 27 (6), 534–548. 10.1080/1354750X.2022.2067233. [DOI] [PubMed] [Google Scholar]
- Dhar R.; Mukherjee S.; Mukerjee N.; Mukherjee D.; Devi A.; Ashraf G. M.; Alserihi R. F.; Tayeb H. H.; Hashem A. M.; Alexiou A.; Thorate N. Interrelation between extracellular vesicles miRNAs with chronic lung diseases. J. Cell Physiol. 2022, 237 (11), 4021–4036. 10.1002/jcp.30867. [DOI] [PubMed] [Google Scholar]
- Wang J.; Chen D.; Ho E. A. Challenges in the development and establishment of exosome-based drug delivery systems. J. Controlled Release 2021, 329, 894–906. 10.1016/j.jconrel.2020.10.020. [DOI] [PubMed] [Google Scholar]
- Kim H.; Jang H.; Cho H.; Choi J.; Hwang K. Y.; Choi Y.; Kim S. H.; Yang Y. Recent Advances in Exosome-Based Drug Delivery for Cancer Therapy. Cancers (Basel) 2021, 13 (17), 4435. 10.3390/cancers13174435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S. C.; Kim O. Y.; Yoon C. M.; Choi D. S.; Roh T. Y.; Park J.; Nilsson J.; Lötvall J.; Kim Y. K.; Gho Y. S. Bioinspired exosome-mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Nano 2013, 7 (9), 7698–710. 10.1021/nn402232g. [DOI] [PubMed] [Google Scholar]
- Adriano B.; Cotto N. M.; Chauhan N.; Jaggi M.; Chauhan S. C.; Yallapu M. M. Milk exosomes: Nature’s abundant nanoplatform for theranostic applications. Bioact Mater. 2021, 6 (8), 2479–2490. 10.1016/j.bioactmat.2021.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar R.; Gorai S.; Krishnan A.; Mukherjee D. Why India needs more exosome research for cancer?. Ann. Med. Surg. 2022, 80, 104265. 10.1016/j.amsu.2022.104265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar R.; Mukherjee D.; Mukerjee N.; Devi A.; Dey A.; Ghosh A. Exosome based theranostic approaches in breast cancer, a new answer of Indian breast cancer-associated health crisis - Correspondence. Int. J. Surg 2022, 105, 106886. 10.1016/j.ijsu.2022.106886. [DOI] [PubMed] [Google Scholar]
- Dhar R.; Bhattacharya B.; Mandal D.; Devi A.; Thorat N. D. Exosome-based cancer vaccine: A cutting-edge approach - Correspondence. Int. J. Surg 2022, 108, 106993. 10.1016/j.ijsu.2022.106993. [DOI] [PubMed] [Google Scholar]
- Krishnan A.; Bhattacharya B.; Mandal D.; Dhar R.; Muthu S. Salivary exosomes: A theranostics secret of oral cancer - Correspondence. Int. J. Surg 2022, 108, 106990. 10.1016/j.ijsu.2022.106990. [DOI] [PubMed] [Google Scholar]
- Hoque S.; Dhar R.; Kar R.; Mukherjee S.; Mukherjee D.; Mukerjee N.; Nag S.; Tomar N.; Mallik S. Cancer Stem Cells (CSCs): key player of radiotherapy resistance and its clinical significance. Biomarkers 2022, 1–28. 10.1080/1354750X.2022.2157875. [DOI] [PubMed] [Google Scholar]