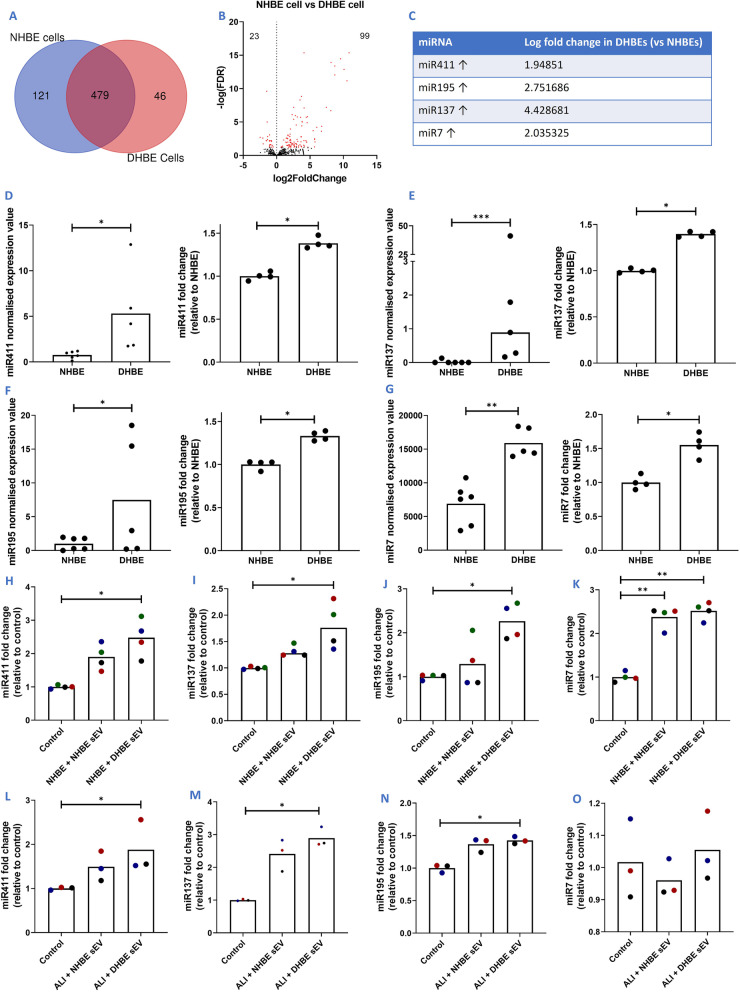
Fig. 6.
Candidate miRNAs are shown to be unregulated in DHBEs and responder cells/cultures co-cultured with diseased sEVs, indicating successful transfer via sEVs. A Overlap of miRNAs detected in NHBE and DHBE cells. B Volcano plot showing differential miRNA expression between NHBE and DHBE cells. C Fold change of candidate miRNA within DHBEs, compared to NHBEs. D–G smRNA-seq expression values and confirmation via qPCR of candidate miRNA: E miR411, E miR137, F miR195, G miR7. H–K Candidate miRNA expression within submerged responder cells co-cultured with NHBE and DHBE-derived sEVs: H miR411, I miR137, J miR195, K miR7. L–O Candidate miRNA expression within healthy ALI cultures co-cultured with NHBE and DHBE derived sEVs: L miR411, M miR137, N miR195, O miR7. Gene normalisation to RNU6B for δδCT and graphs show fold change relative to NHBEs or control of no added sEVs. N = 3-. Each colour represents a different donor and experiment. Significant differences between groups shown ***/red = p or p.adj < 0.001; **p < 0.01; *p < 0.05, Mann–Whitney test and Friedmans test