Figure 2.
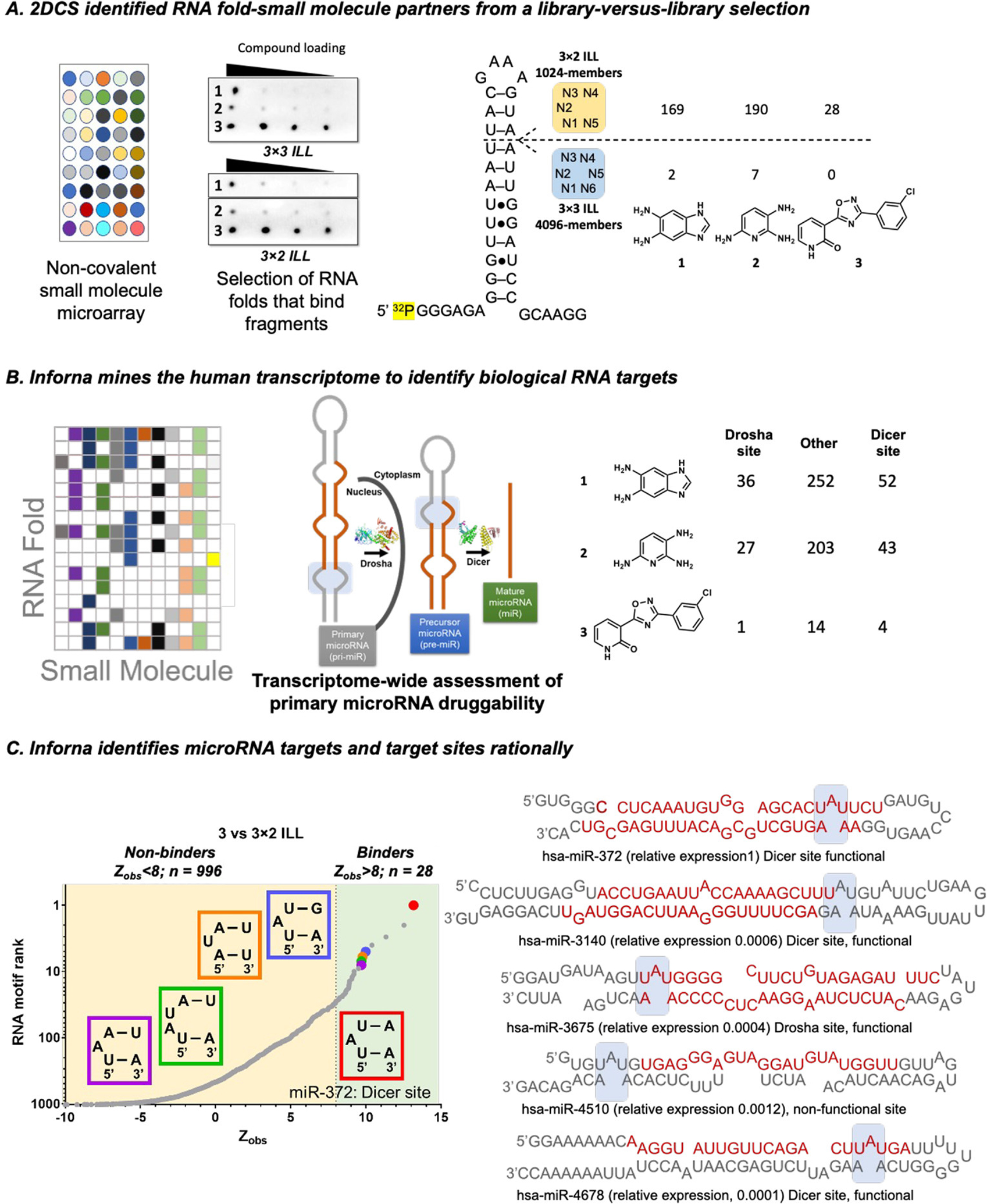
RNA targets of fragments, as selected by 2DCS. (A) The fragments were studied for binding RNA libraries that display 3 × 3 or 3 × 2 nucleotide internal loops (3 × 3 ILL and 3 × 2 ILL, respectively) using AbsorbArray, where the compounds are non-covalently immobilized on agarose coated glass slide. Three fragments (1, 2, and 3) bind RNAs specifically as determined by a secondary selection in the presence of competitor oligonucleotides (2DCS) (left). RNA targets of compounds were identified from sequencing analysis. The number of RNA targets yielded for each compound for individual RNA ILLs when a cutoff of Zobs > 8 was employed (right). (B) Scheme for RNA mining using Inforna. Inforna mines all human miRNA precursors and identifies the RNA target that has a small molecule-binding motif present in its secondary structure. (C) Inforna mining further studies whether the RNA motif is present in a functional (Dicer or Drosha) site or non-functional site of the pri- and pre-miRNAs. Fragment 3 is predicted to bind 5′UAU/3′A_A with the highest Zobs (left). Secondary structures of all miRNAs containing the A bulge (5′UAU/3′A_A) (right). Among them, only four miRNAs (miR-372, miR-3140, miR-3675, and miR-4678) harbor the A bulge in a functional site. However, the expression of miR-3140, miR-3675, and miR-4678 is >100-fold lower than miR-372 in AGS cells.
