Extended Data Fig. 2 |. Superposition of the cryo-EM map densities of the PSM, PΦB, and the bilin-binding GAF domain pocket with the X-ray crystallographic model of the PSM.
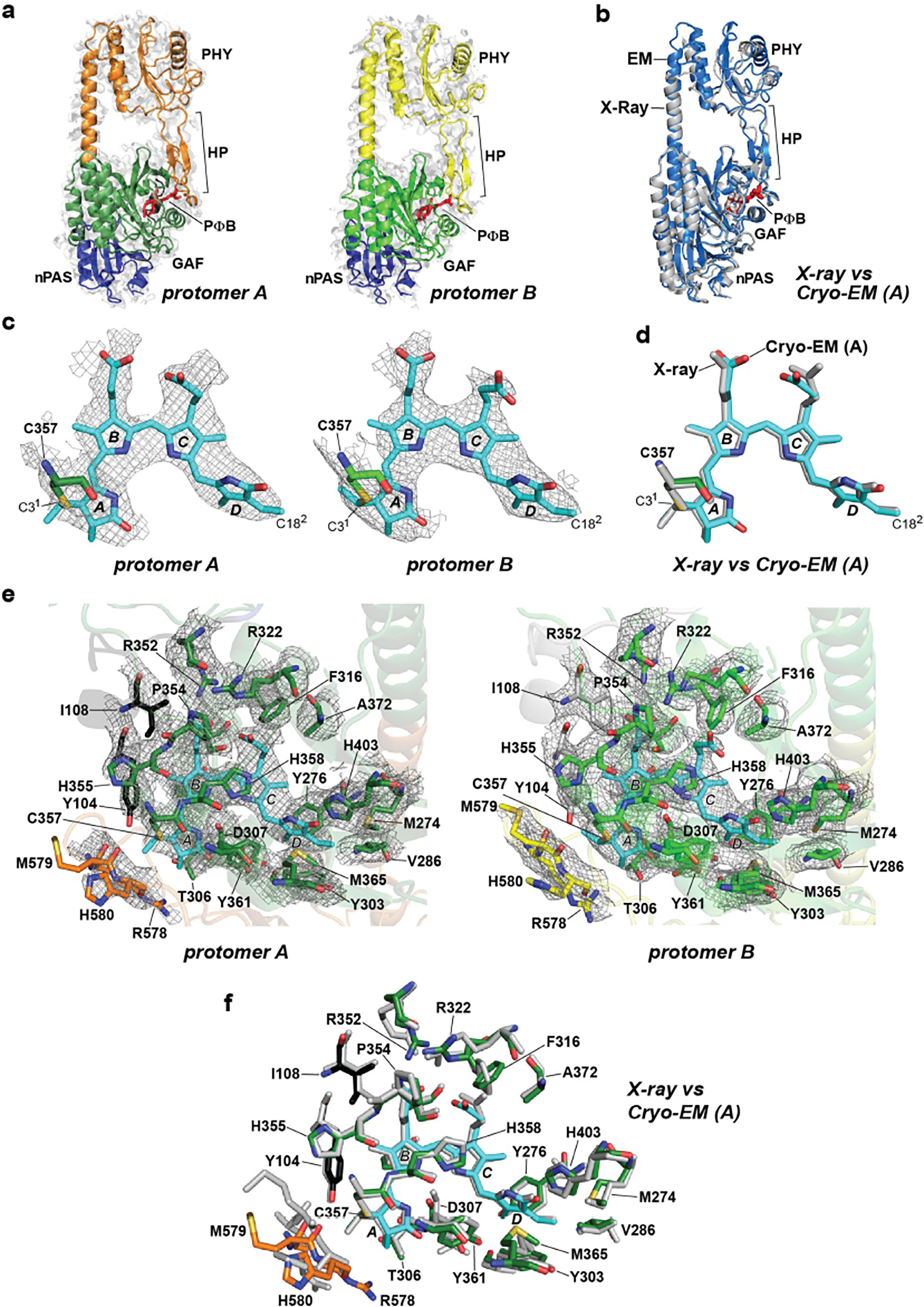
Motifs/residues are colored as in Fig. 1b. a, Fitting of the nPAS, GAF and PHY domains, and the hairpin (HP) motif within the EM map density (light grey surface) of protomer A and protomer B. PΦB is shown in red. b, Superposition of the PhyB PSM determined by cryo-EM of the full-length PhyB (protomer A; slate blue) and by X-ray crystallography of the PhyB PSM (grey; residues 90–624, PDB ID code, 4OUR32). c, PΦB conformations (in sticks) in protomers A and B modeled within the EM map density (grey mesh). The A-D pyrrole rings are labeled along with Cys357 that forms a thioether linkage to the C31 carbon of PΦB. The D ring C182 carbon is indicated. d, Superposition of the PΦB structures determined by cryo-EM of the full-length PhyB protomer A (colored) and by X-ray crystallography of the PhyB PSM (grey). e, The bilin-binding pockets of protomers A and B highlighting neighboring amino acids (sticks) and superposed in the EM map density (grey mesh). f, Superposition of the bilin-binding pocket determined from the cryo-EM structure of full-length PhyB protomer A with the X-ray crystallographic structure of the PhyB PSM.
