ABSTRACT
Background
Patients hospitalized with COVID-19 are at significant risk for superimposed bacterial pneumonia. However, diagnosing superinfection is challenging due to its clinical resemblance to severe COVID-19. We therefore evaluated whether the immune biomarker, procalcitonin, could facilitate the diagnosis of bacterial superinfection.
Methods
We retrospectively identified 185 patients hospitalized with severe COVID-19 who underwent lower respiratory culture; 85 had evidence of bacterial superinfection. Receiver operating characteristic curve and area under the curve (AUC) analyses were performed to assess the utility of procalcitonin for diagnosing superinfection.
Results
This approach demonstrated that procalcitonin measured at the time of culture was incapable of distinguishing patients with bacterial infection (AUC, 0.52). The AUC not affected by exposure to antibiotics, treatment with immunomodulatory agents, or timing of procalcitonin measurement.
Conclusion
Static measurement of procalcitonin does not aid in the diagnosis of superinfection in severe COVID-19.
KEYWORDS: COVID-19, procalcitonin, critical care, bacterial superinfection, pneumonia, biomarker
Introduction
Bacterial pneumonia is an important complication in patients hospitalized with COVID-19. Its incidence approaches 45% in those receiving mechanical ventilation, and it is associated with increased 28-day mortality [1]. However, diagnosing bacterial superinfection is challenging due to its overlapping clinical features with severe COVID-19, including fever, hypoxemia, and radiographic infiltrates. As a result, patients with pure viral infection are often exposed to unnecessary empiric antibiotics, especially in the intensive care unit (ICU). It is therefore essential to identify reliable diagnostics for superinfection to guide antimicrobial stewardship in patients with COVID-19.
Procalcitonin (PCT) is an inflammatory biomarker that has been used as an adjunctive test for bacterial pneumonia for over 20 years. The role of PCT in identifying superinfection during COVID-19 remains equivocal. May et al. and Relph et al. have demonstrated limited utility [2,3], while others have argued for its efficacy [4–6]. Importantly, these studies were limited mostly to patients with coinfection upon presentation to the hospital, which is rare compared to secondary bacterial pneumonia (defined by Russell et al. as occurring > 48 h after admission) [7]. In addition, the reported cohorts included patients with varying disease severity, which may confound the interpretation of PCT [8]. In line with this hypothesis, we previously demonstrated that PCT levels are associated with clinical severity in non-COVID respiratory viral infection [9]. Therefore, we sought to evaluate the diagnostic utility of PCT for bacterial pneumonia in COVID-19 – specifically in patients with severe disease after admission to the hospital.
Materials and methods
We performed a retrospective cohort study of adult patients admitted to Yale New Haven Hospital between 3 January 2020 and 1 July 2020 with severe COVID-19, as defined by the National Institute of Health COVID-19 Treatment Guidelines: positive SARS-CoV-2 nucleic acid amplification testing and hypoxemia (SpO ≤ 94% and/or use of supplemental oxygen). We subsequently identified the subgroup of 185 patients from whom lower respiratory culture (LRCx) was collected. Of the 185 LRCx, 106 were sputum samples and 79 were tracheal aspirates. If multiple specimens were obtained, the index culture was used for analysis. Patients with positive fungal LRCx and those with positive blood or urine cultures were excluded to restrict the analysis to bacterial respiratory infection.
Data were collected by Yale’s Joint Data Analytic Team and verified via chart review. Unless otherwise indicated, lab values used for analysis (e.g. PCT, creatinine, etc.) were those closest to the time of LRCx. If no lab value was recorded within 36 hours of LRCx, it was recorded as missing. Data were missing at a rate of less than 15% for all variables. All patients had a PCT measured within 36 h of LRCx. Statistical analysis was conducted using R (V.3.6.0, Austria). To determine significance (defined as p < 0.05), the Chi-squared or Kruskal-Wallis test was performed due to non-normal data distributions. To assess correlations between biomarkers, Spearman’s rank correlation coefficient was utilized due to the likelihood of non-linear relationships between values. The study was approved by Yale’s institutional review board (approval #2000023067). Informed consent was not required due to the non-interventional study design.
Results
We identified 1308 patients hospitalized with COVID-19 during the study period. Of these, 185 had both severe COVID-19 and a LRCx; 100 had no bacterial growth and 85 had culture-proven superinfection. As shown in Table 1, these groups were well-matched in terms of demographic and clinical variables. Outside of mechanical ventilation, the groups demonstrated similar disease severity as indicated by organ failure, ICU admission, and mortality. Importantly, > 70% of LRCx were obtained after 48 h of hospitalization, thus focusing the analysis on secondary bacterial pneumonia (median PCT was 95.3 h after admission). The distribution of bacterial etiologies showed a predominance of Staphylococcus aureus (34.7%) and Pseudomonas aeruginosa (12.2%), consistent with prior reports [2,7].
Table 1.
Demographic, clinical, and laboratory characteristics of COVID-19 patients with bacterial superinfection compared to those with negative lower respiratory culture (LRCx).
| Variables shown as median (IQR) except where otherwise specified | Pure viral infection (n = 100) |
Bacterial superinfection (n = 85) |
p-value |
|---|---|---|---|
| Age, median (IQR) | 64.50 (55.00, 76.25) | 63.00 (49.00, 69.00) | 0.128 |
| Female sex, n (%) | 34 (34.0%) | 23 (27.1%) | 0.308 |
| Body mass index | 28.22 (23.92, 35.78) | 30.28 (25.63, 36.14) | 0.353 |
| ICU admission, n (%) | 84 (84.0%) | 75 (88.2%) | 0.409 |
| Mechanical ventilation, n (%) | 57 (57.0%) | 68 (80.0%) | <0.001 |
| Renal failure, n (%) | 67 (67.0%) | 65 (76.5%) | 0.156 |
| Shock, n (%) | 73 (73.0%) | 66 (79.5%) | 0.304 |
| Mortality, n (%) | 22 (22.0%) | 28 (32.9%) | 0.095 |
| Steroids within 24 h of LRCx, n (%) | 22 (22.0%) | 23 (27.1%) | 0.424 |
| Tocilizumab within 14d of LRCx, n (%) | 67 (67.0%) | 47 (55.3%) | 0.103 |
| Biomarkers at time of LRCx | |||
| PCT | 0.40 (0.13, 0.76) | 0.30 (0.14, 0.88) | >0.999* |
| WBC | 8.30 (4.80, 13.60) | 10.20 (6.00, 14.10) | >0.999* |
| Creatinine | 1.23 (0.82, 2.48) | 1.06 (0.71, 1.79) | >0.999* |
| C-reactive protein | 91.85 (30.08, 153.47) | 49.75 (15.05, 108.25) | 0.289 |
| D-Dimer | 2.02 (1.13, 5.00) | 2.59 (0.93, 4.61) | >0.999* |
| Ferritin | 1143.00 (653.50, 1982.25) | 816.00 (332.50, 1689.75) | 0.210 |
| Temperature | 98.20 (97.80, 99.23) | 98.90 (98.00, 99.75) | 0.156 |
| Microbiological data | Count | ||
| Staphylococcus aureus, n (%) | 34 (34.7%) | ||
| Methicillin-resistant S. aureus, n (%) | 6 (6.1%) | ||
| Pseudomonas aeruginosa, n (%) | 12 (12.2%) | ||
| Haemophilus influenzae, n (%) | 8 (8.2%) | ||
| Klebsiella pneumoniae, n (%) | 7 (7.1%) | ||
| Enterobacter spp, n (%) | 7 (7.1%) | ||
| Escherichia coli, n (%) | 6 (6.1%) | ||
| Beta-hemolytic streptococcus, n (%) | 5 (5.1%) | ||
| Moraxella catarrhalis, n (%) | 4 (4.1%) | ||
| Other bacterial pathogens†, n (%) | 9 (9.2%) | ||
| Polymicrobial infection, n (%) | 13 (15.3%) |
*Bonferroni-adjusted p-values are reported for biomarkers to adjust for multiple comparisons.
†Other bacterial pathogens include species observed < 3 times. These include Streptococcus pneumoniae (2), Serratia marcescens (2), Stenotrophomonas maltophilia (2), Citrobacter koseri (1), Morganella morganii (1), Proteus mirabilis (1).
p-values were calculated via chi-squared test or Kruskal-Wallis test, as appropriate.
Abbreviations: intensive care unit (ICU), lower respiratory culture (LRCx), interquartile range (IQR), procalcitonin (PCT), white blood cell count (WBC).
PCT was similar in patients with pure viral infection compared to those with bacterial superinfection (Figure 1a). The area under the ROC curve (AUROC) for identifying bacterial pneumonia was 0.52 (95% CI: 0.43–0.60), indicating that PCT is an ineffective test for superinfection in severe COVID-19 (Figure 1b). Importantly, we noted that 63% of patients had received antibiotics within 24 hours of LRCx. To determine whether antibiotic exposure influenced the diagnostic utility of PCT, we evaluated the AUROC in exposed vs unexposed patients. This analysis revealed similarly poor performance in both subgroups: AUROCexposed = 0.49; AUROCunexposed = 0.50. In addition, we considered whether treatment with tocilizumab (an anti-IL-6 antibody) could have influenced PCT expression due to its anti-inflammatory effects. However, in patients with bacterial superinfection, PCT levels were not significantly affected by tocilizumab exposure (median PCTexposed = 0.30 ng/mL; median PCTunexposed = 0.31 ng/mL; p = 0.53).
Figure 1.
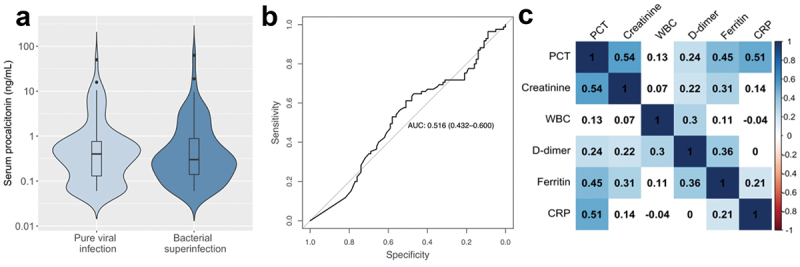
Serum procalcitonin as a biomarker of bacterial superinfection in patients with severe COVID-19. a, Procalcitonin levels in patients with pure COVID-19 infection and those with bacterial superinfection. b, Receiver operating characteristic curve defining the utility of PCT in diagnosing superinfection. c, Spearman’s rank correlation between biomarkers at the time of LRCx.
Using a cutoff of 0.5 ng/mL, the sensitivity and specificity of PCT were unacceptably low at 33% and 60%, respectively. At a threshold of 0.25 ng/mL, the negative predictive value (NPV) was similarly poor (51%). In contrast, we found that PCT was more effective at predicting severe disease within the study population, as indicated by the AUROC for shock (0.65), renal failure (0.72), mechanical ventilation (0.61), ICU admission (0.64), and death (0.62). In addition, PCT correlated significantly with several non-specific markers of inflammation including ferritin (Spearman’s ρ = 0.45, p < 0.001) and CRP (ρ = 0.51, p < 0.001) (Figure 1c). We then performed a sub-analysis restricted to patients with LRCx > 48 h after admission; i.e. those who developed bacterial superinfection during hospitalization [7]. In this subgroup, AUROC was similarly poor (0.51), and sensitivity and specificity at a cutoff of 0.5 ng/mL were similarly low (28% and 63% respectively).
Discussion
Taken together, our results suggest that PCT is a poor biomarker for bacterial superinfection in severe COVID-19. This conclusion diverges from prior reports supporting the utility of PCT [4–6]; the difference is likely explained by our use of study groups with similar clinical severity, which minimized the effect of this potential confounder. In addition, we found that PCT had a poor NPV (51%), in contrast to the excellent NPV reported by So et al. (>90%) at the same cutoff of 0.25 ng/mL [10]. This discrepancy is likely a result of restricting our analysis to patients with severe disease and therefore higher baseline PCT. In this population with high pre-test probability, which presents the most significant diagnostic challenge to clinicians, NPV was markedly lower.
The conclusions presented here are supported by the work of Pickens et al, who also demonstrated comparable PCT levels in pure viral infection and bacterial pneumonia [1]. Our study extends these findings, as we applied ROC analysis to formally assess the biomarker’s diagnostic performance. In addition, our conclusions align with those of May et al. and Relph et al. who argued against using PCT for identifying bacterial coinfection upon hospital admission [2]. However, our study focuses on patients with hospital-acquired secondary bacterial pneumonia, which is significantly more common than community-acquired coinfection [7].
Rather than indicating bacterial pneumonia, PCT appears to reflect the severity of lung inflammation, consistent with prior studies (Figure 1c) [8]. Thus, we propose that PCT be interpreted as a general indicator of the inflammatory host response, which may derive from superinfection or severe COVID-19 itself.
It is important to note that patients were identified retrospectively in this study according to receipt of LRCx; respiratory cultures were not obtained in all patients. Consequently, these data should not be used to estimate the incidence of bacterial superinfection in our cohort. In addition, the study was restricted to patients with severe disease, in whom baseline PCT is often elevated [8]. As such, it was not designed to assess the utility of low PCT in ruling out bacterial pneumonia during mild COVID-19. Although evaluation of PCT kinetics may aid in the detection of nosocomial superinfection, the frequency of PCT values in our dataset precluded our examination of this question. A final point of consideration is that our study was conducted early in the pandemic, prior to emergence of the Omicron variant and availability of vaccines, both of which have contributed to a shift towards milder disease.
In the absence of specific clinical, radiographic, and biochemical tests for bacterial pneumonia in COVID-19, direct microbiologic sampling may represent the optimal means of identifying this important complication and improving antibiotic stewardship in hospitalized patients [1]. Ongoing evaluation of the safety and efficacy of routine LRCx for the diagnosis of superinfection will be needed.
Funding Statement
The work was supported by the Cystic Fibrosis Foundation [GAUTAM20D0]; U.S. Department of Defense [PR181442]; NHLBI [HL007778, HL154641, HL159422]; NHLBI [AI089992-09S2]; NIAID [AI007517]; Doris Duke COVID-19 FRCS at Yale [2021266]; Parker B. Francis Fellowship; Claude D. Pepper Older Americans Independence Center at Yale [P30AG021342]; Robert E. Leet and Clara Guthrie Patterson Trust; Yale Center for Clinical Investigation Scholar Award; U.S. Department of Veterans Affairs Merit Award [BX004661]
Abbreviations
- BMI
Body mass index
- ICU
Intensive care unit
- IQR
Interquartile range
- LRCx
Lower respiratory culture
- PCT
Procalcitonin
- SpO2
Oxygen saturation
- WBC
White blood cell count
Disclosure statement
The authors declare no relevant conflicts of interest. SG has been a consultant to AstraZeneca.
References
- [1].Pickens CO, Gao CA, Cuttica MJ, et al. Bacterial superinfection pneumonia in patients mechanically ventilated for COVID-19 pneumonia. Am J Respir Crit Care Med. 2021;204(8):921–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].May M, Chang M, Dietz D, et al. Limited utility of procalcitonin in identifying community-associated bacterial infections in patients presenting with coronavirus disease 2019. Antimicrob Agents Chemother. 2021;65(4). DOI: 10.1128/AAC.02167-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Relph KA, Russell CD, Fairfield CJ, et al. Procalcitonin is not a reliable biomarker of bacterial coinfection in people with coronavirus disease 2019 undergoing microbiological investigation at the time of hospital admission. Open Forum Infect Dis. 2022;9(5). DOI: 10.1093/ofid/ofac179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pink I, Raupach D, Fuge J, et al. C-reactive protein and procalcitonin for antimicrobial stewardship in COVID-19. Infection. 2021;49(5):935–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hughes S, Mughal N, Moore LSP.. Procalcitonin to guide antibacterial prescribing in patients hospitalised with covid-19. Antibiotics. 2021;10(9):1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Atallah NJ, Warren HM, Roberts MB, et al. Baseline procalcitonin as a predictor of bacterial infection and clinical outcomes in COVID-19: a case-control study. PLoS ONE. 2022;17(1):e0262342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Russell CD, Fairfield CJ, Drake TM, et al. Co-infections, secondary infections, and antimicrobial use in patients hospitalised with COVID-19 during the first pandemic wave from the ISARIC WHO CCP-UK study: a multicentre, prospective cohort study. Lancet Microbe. 2021;2(8):e354–365. DOI: 10.1016/S2666-5247(21)00090-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shen Y, Cheng C, Zheng X, et al. Elevated procalcitonin is positively associated with the severity of COVID-19: a meta-analysis based on 10 cohort studies. Medicina (Kaunas). 2021;57(6):594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gautam S, Cohen AJ, Stahl Y, et al. Severe respiratory viral infection induces procalcitonin in the absence of bacterial pneumonia. Thorax. 2020;75(11):974–981. [DOI] [PubMed] [Google Scholar]
- [10].So W, Simon MS, Choi JJ, et al. Characteristics of procalcitonin in hospitalized COVID-19 patients and clinical outcomes of antibiotic use stratified by procalcitonin levels. Intern Emerg Med. 2022;17(5):1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]


