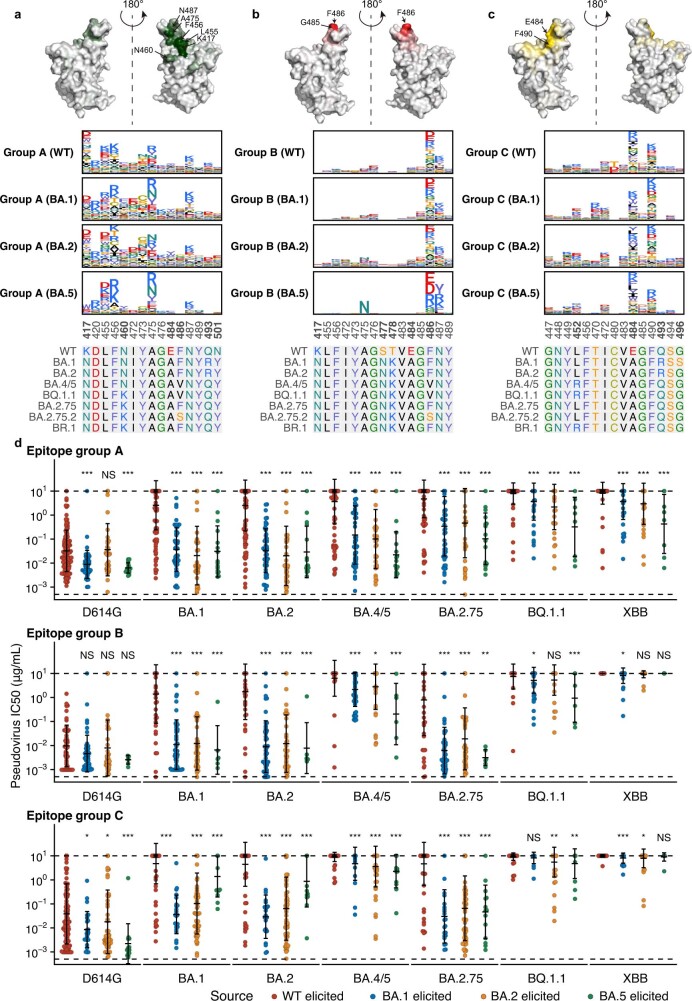
Extended Data Fig. 6. Escape hotspots and neutralization of mAbs in epitope groups A, B and C.
a-c, Average escape scores from DMS of epitope groups A (a), B (b), C (c) and each RBD residue. Scores are projected onto the structure of SARS-CoV-2 RBD (PDB: 6M0J). Average escape maps that indicate the score of each mutation from DMS on escape hotspots of antibodies, grouped by their sources, in epitope groups A (a), B (b) and C (c), and corresponding sequence alignment of SARS-CoV-2 WT and Omicron RBDs are also shown. The height of each amino acid in the escape maps represents its mutation escape score. Mutated sites in Omicron variants are marked in bold. d, Pseudovirus-neutralizing IC50 of antibodies in group A, B, and C, from wild-type vaccinated or convalescent individuals (WT-elicited, n = 133, 50, 106 for A-C, respectively), BA.1 (BA.1-elicited, n = 51, 49, 24), BA.2 (BA.2-elicited, n = 34, 36, 56) and BA.5 convalescent individuals (BA.5-elicited, n = 16, 6, 14). The geometric mean IC50s are labelled, and error bars indicate the geometric standard deviation. P-values are calculated using two-tailed Wilcoxon rank sum tests. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, not significant, p > 0.05. All neutralization assays were conducted in at least two independent experiments.