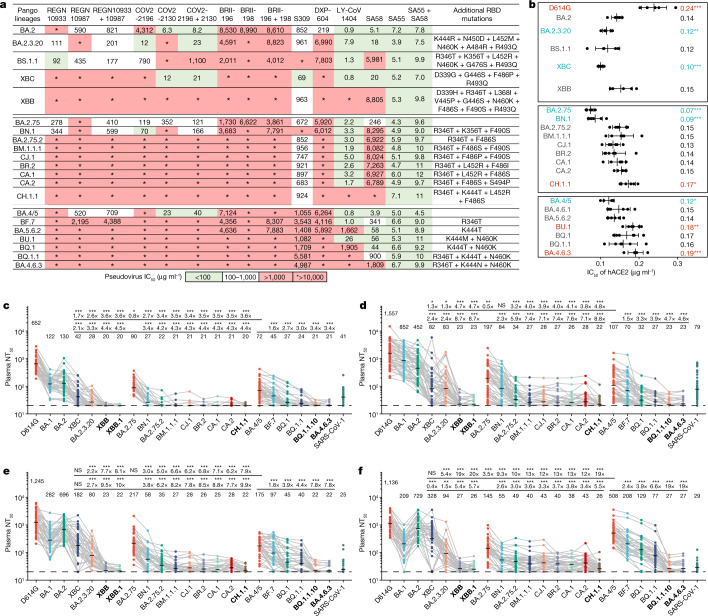
Fig. 2. Convergent Omicron subvariants induce NAb evasion.
a, IC50 of therapeutic NAbs against VSV-based pseudoviruses with spike glycoproteins of emerging SARS-CoV-2 BA.2, BA.5 or BA.2.75 convergent subvariants. b, Relative hACE2-binding capability measured by IC50 of hACE2 against pseudoviruses of variants. Error bars indicate mean ± s.d. of n = 5 biologically independent replicates. P values were calculated using two-tailed Student’s t-test. *P < 0.05, **P < 0.01 and ***P < 0.001. There is no label on variants with P > 0.05. Variants with significantly stronger binding are coloured blue, whereas those with weaker binding are coloured red. c–f, Pseudovirus-neutralizing titres against SARS-CoV-2 D614G and Omicron subvariants of plasma from vaccinated or convalescent individuals of breakthrough infection. Individuals who had received three doses of CoronaVac (n = 40) (c), individuals who had been infected with BA.1 after receiving three doses of CoronaVac (n = 50) (d), individuals who had been infected with BA.2 after receiving three doses of CoronaVac (n = 39) (e) and individuals who had been infected with BA.5 after receiving three doses of CoronaVac (n = 36) (f) are shown. The geometric mean titres are labelled. Statistical tests were performed using two-tailed Wilcoxon signed-rank tests of paired samples. *P < 0.05, **P < 0.01, ***P < 0.001 and not significant (NS) P > 0.05. NT50 against BA.2-derived and BA.2.75-derived subvariants were compared with that against BA.4/5 (the upper line) and BA.2.75 (the lower line); BA.4/5-derived subvariants were only compared with BA.4/5. Dashed lines indicate the limit of detection (LOD, NT50 = 20). Strains showing the strongest evasion are in bold. All neutralization assays were conducted in at least two independent experiments.