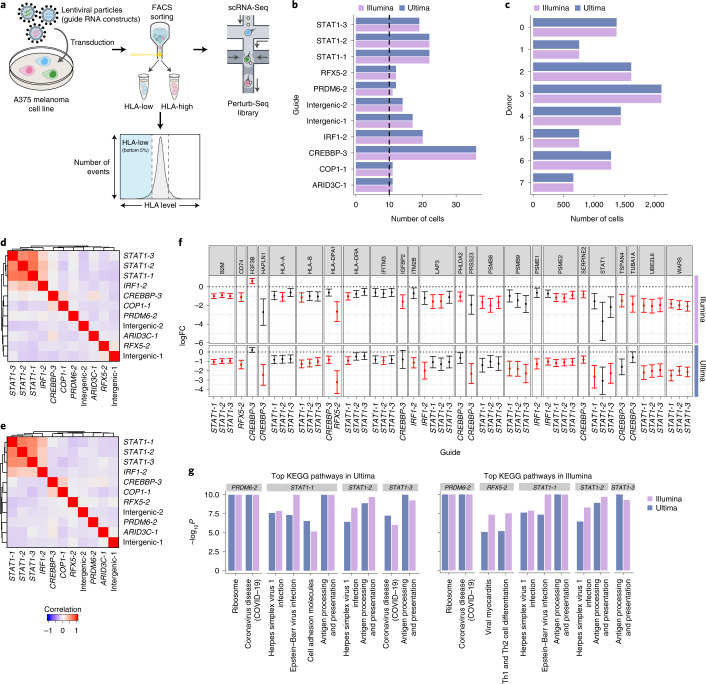
Fig. 4. Perturb-Seq.
a, Perturb-Seq to find regulators of MHC Class I in melanoma. We transduced A375 melanoma cells with a genome-wide library, and cells with low HLA expression were enriched by flow cytometry before scRNA-seq. b, Number of cells with each perturbation, only plotting those with >10 cells, excluding non-targeting and background guides. c, Number of cells with each Cell Hashing label. d,e, Guide similarity heat maps in Ultima (d) and Illumina (e). Effects of each guide on each gene in the Illumina data were calculated with an elastic-net-based approach as in MIMOSCA. The matrix of guides by genes with (uncorrected) P < 0.05 was extracted; the correlation between guides was calculated and plotted as a heat map. f, We extracted all gene–guide pairs from our DE analysis with FDR < 0.05 in either Illumina or Ultima. For each of these guide–gene pairs, we plotted the logFC on the y-axis (with 95% confidence intervals) and guide on the x-axis, with each box being a different gene, for both Illumina and Ultima. We included all guides that targeted a gene with any significantly different guides. The dots are colored red if significant (FDR < 0.05) and black if not. g, KEGG enrichment analysis for each guide. The ten pathways with smallest P values in both Illumina (left) and Ultima (right) are plotted as their −log10P in both Illumina and Ultima. All pathways were significant in both Illumina and Ultima at FDR of 0.05.