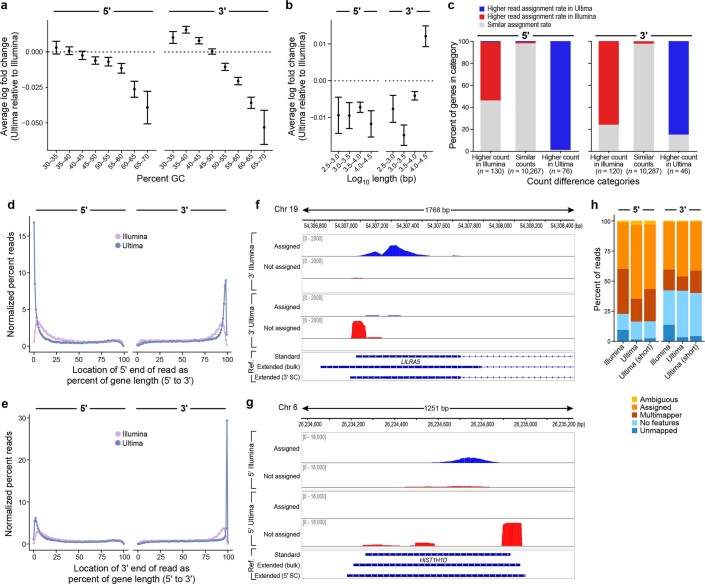
Extended Data Fig. 4. Sequencing biases in matched 5′ and 3′ PBMC libraries.
(a) Genes were assigned to bins by GC content (exonic sequences only) and the average logFC for each bin between Ultima and Illumina is shown with 95% CI. Positive logFC indicates higher expression in Ultima. (b) Genes were assigned to bins by log10 length (total number of bp in at least one exon for a given gene) and the average logFC for each bin between Ultima and Illumina is shown with 95% CI. Positive logFC indicates higher expression in Ultima. (c) Differences in read assignment ratio for genes categorized based on their expression being similar or higher in one platform than the other. Number of genes in each category shown on top of each bar. See Methods for additional details. (d) For each library (5′ PBMC, left, and 3′PBMC, right), the 5′ end of each read is shown at relative positions along the length of each gene. (e) Relative position of the 3′ end of reads as in (d). (f) LILRA5 sequence coverage of 3′ libraries with Illumina (top two tracks) and Ultima (next two tracks). Standard gene annotation with introns as lines, exons as boxes, and arrows for direction of transcription shown in bottom track. Extended reference annotation using bulk or single cell (SC) Ultima data shown at bottom. Reads assigned (blue) or not (red) to LILRA5 by Cell Ranger. (g) HIST1H1D sequence coverage of 5′ libraries shown as in (f). (h) Bar plots showing classification of reads by FeatureCount. Ultima (short) reads had Read 2 trimmed to 45 bases for 5′ and 55 bases for 3′ to match Illumina read lengths. Assigned: assigned to genes (uniquely mapped reads that overlap a gene; not all of these are assigned to a gene by Cell Ranger, which only assigns reads that completely overlap a gene); ambiguous: uniquely mapped reads that overlap the exons of multiple genes; no feature: uniquely mapped reads that do not overlap any exons; multimappers: map to multiple genomic locations (note Cell Ranger is able to recover multimappers if they only overlap one gene); unmapped: map to no genomic locations. Each read can only fit in one category. Ultima data sampled to have the same number of UMIs ((a) and (b)) or reads (other panels) as the Illumina data.