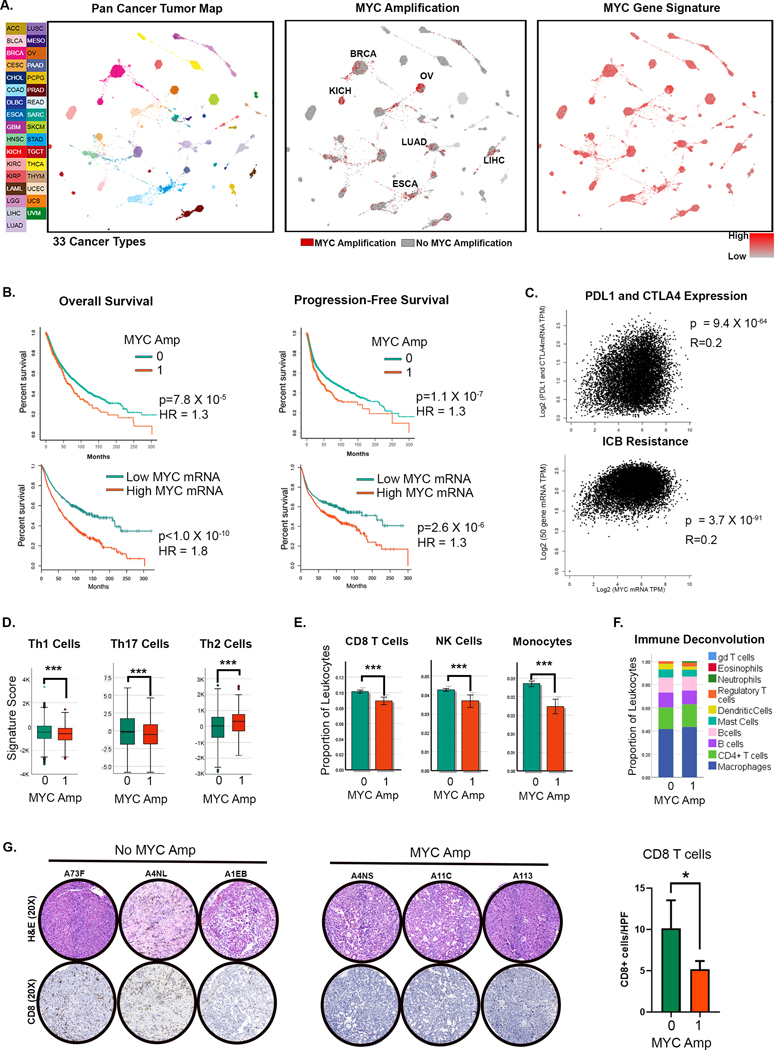
Figure 1. The MYC Oncogene Drives Human Tumorigenesis Associated with Immune Evasion.
A. TumorMap rendering of the Pan-Cancer-Atlas of 33 cancers; tumors are defined by different tissues of origin in the first box. Second box shows the prevalence of MYC genomic amplification. Epithelial tumors with high rates of MYC amplification are annotated. Third box shows the gene expression of a signature derived from MYC amplification. (LAML- Acute Myeloid Leukemia, ACC- Adrenocortical carcinoma, BLCA- Bladder Urothelial Carcinoma, LGG- Brain Lower Grade Glioma, BRCA- Breast invasive carcinoma, CESC- Cervical squamous cell carcinoma and endocervical adenocarcinoma, CHOL- Cholangiocarcinoma, LCML- Chronic Myelogenous Leukemia, COAD- Colon adenocarcinoma, ESCA- Esophageal carcinoma, GBM- Glioblastoma multiforme, HNSC- Head and Neck squamous cell carcinoma, KICH- Kidney Chromophobe, KIRC- Kidney renal clear cell carcinoma, KIRP- Kidney renal papillary cell carcinoma, LIHC- Liver hepatocellular carcinoma, LUAD- Lung adenocarcinoma, LUSC- Lung squamous cell carcinoma, DLBC- Lymphoid Neoplasm Diffuse Large B-cell Lymphoma, MESO- Mesothelioma, OV- Ovarian serous cystadenocarcinoma, PAAD- Pancreatic adenocarcinoma, PCPG- Pheochromocytoma and Paraganglioma, PRAD- Prostate adenocarcinoma, READ- Rectum adenocarcinoma, SARC- Sarcoma, SKCM- Skin Cutaneous Melanoma, STAD- Stomach adenocarcinoma, TGCT- Testicular Germ Cell Tumors, THYM- Thymoma, THCA- Thyroid carcinoma, UCS- Uterine Carcinosarcoma, UCEC- Uterine Corpus Endometrial Carcinoma, UVM- Uveal Melanoma).
B. Overall and progression-free survival in patients with MYC amplification and MYC overexpression (top and bottom quartiles).
C. Correlation of MYC expression with gene expression of immune checkpoints PDL1 and CTLA4 (top) and with gene signature of immune checkpoint blockade (ICB) (Jiang et al. 2018 (28); bottom).
D. Expression of gene signatures associated with T helper cells 1 (Th1), Th17 and Th2 compared between tumors with (1) and without (0) MYC amplification.
E. Expression of gene signatures associated with CD8 T cells, NK cells and monocytes compared between tumors with (1) and without (0) MYC amplification.
F. Immune deconvolution analysis showing comparison of abundance of different immune subsets between tumors with (1) and without (0) MYC amplification.
G. Immunohistochemistry showing CD8 T-cell infiltration in representative HCC tumors from the TCGA cohort with (1, n=20) and without MYC amplification (0, n=8). *p<0.05, ***p<0.0001.