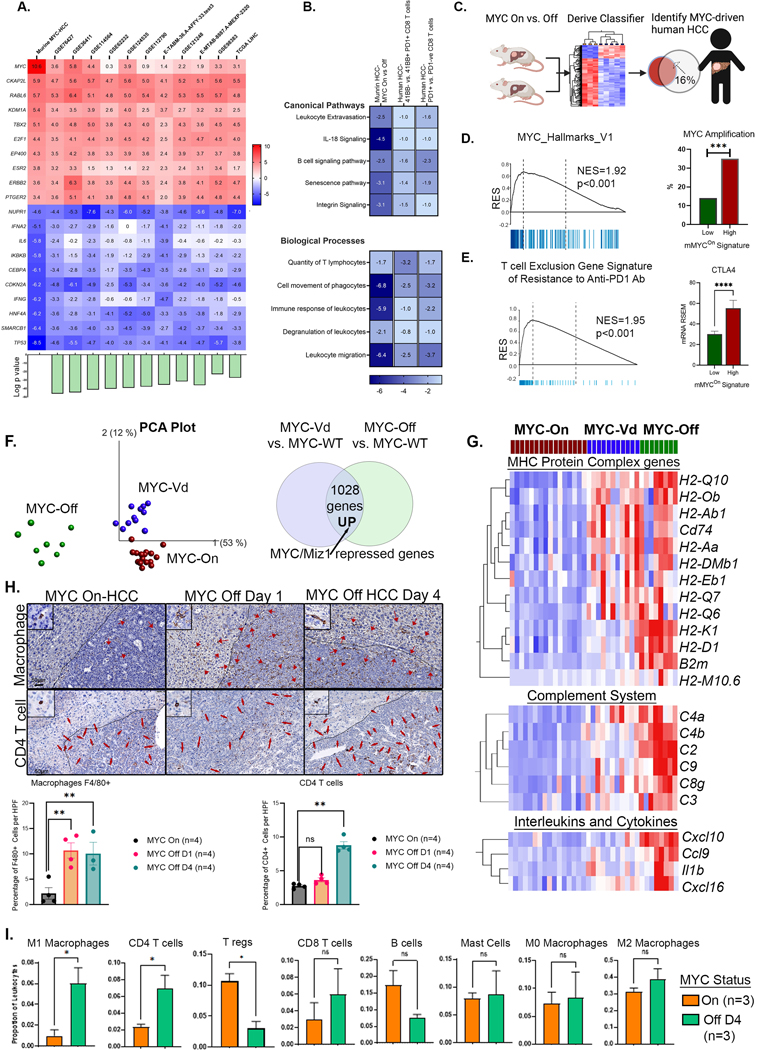
Figure 2: MYC causes Reversible Transcriptional Repression of Antitumor Immune Responses.
A. Heat map shows similarity between MYC-driven murine HCC (MYC-On, n=3 vs. MYC-Off, n=3) and 11 human HCC transcriptome data sets including the TCGA cohort; the top 10 upregulated and downregulated genes with enrichment z-scores across the different data sets are shown. Bar plot at the bottom shows log of p-value of similarity between the individual cohorts and murine MYC-HCC.
B. Heatmap shows z-scores of enrichment of canonical pathways and biological functions in murine MYC-HCC transcriptome and the human HCC immune signatures of exhausted PD1+ CD8 T cells(31) and inactivated 41BB-ve PD1+ T cells in human HCC(30).
C. Gene signature derived from murine MYC HCC used to classify human HCC in the TCGA cohort to identify MYC-driven human HCCs (created using Biorender.com).
D. Human HCCs enriched in mMYCOn signature show MYC molecular pathway activation and strong association with MYC gene amplification.
E. Human HCCs with high expression of mMYCOn signature showed enrichment of a gene program of T cell exclusion and resistance to anti-PD1 checkpoint therapy (32), and upregulation of CTLA4.
F. PCA plot comparing the transcriptional profile of a transgenic model of HCC overexpressing wild-type MYC (MYCOn, n=16), a mutant MYC defective in Miz1 binding (V394D) (MYCVd, n=11) and tumors upon MYC inactivation for a short time point of 16 hours (MYCOff, n=8)(36) to identify MYC/Miz-1 repressed genes.
G. Heatmap shows genes which are reversibly repressed by MYC in the MHC protein complex, complement system and cytokines.
H. Immunohistochemistry shows the abundance of macrophages (F4/80+) by day 1 and CD4 T-cell recruitment by day 4 of MYC inactivation in HCC.
I. Immune deconvolution analysis showing comparison of abundance of different immune subsets in murine MYC HCC upon MYC inactivation.
**p<0.01, ***p<0.0001.