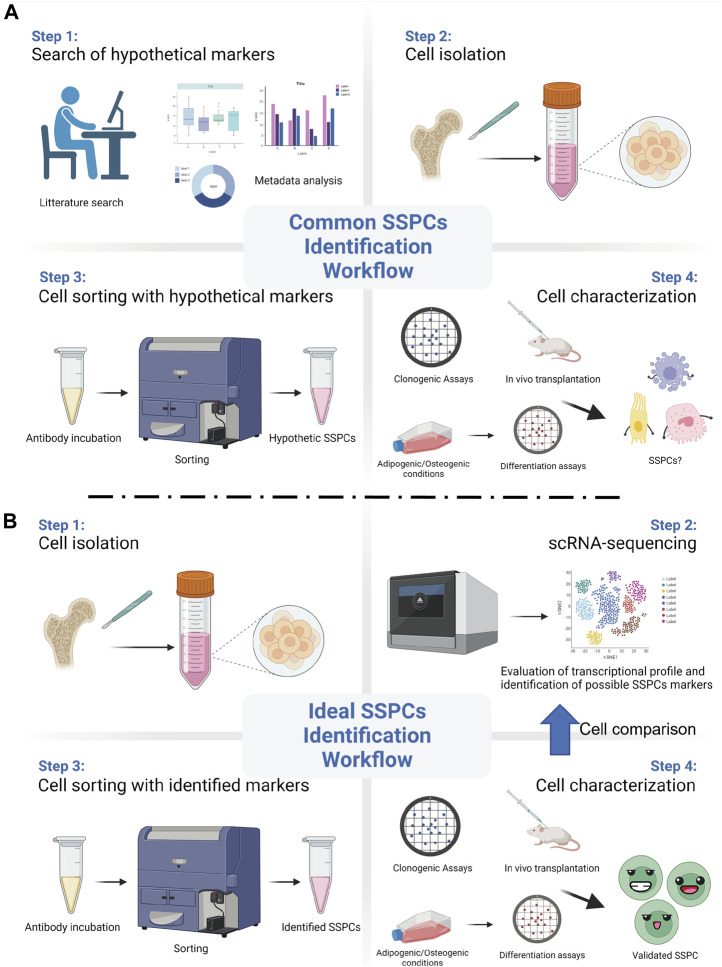
FIGURE 1.
Common vs ideal SSPCs identification workflow. (A) A common and biased approach starts with the selection of SSPCs markers using previously published data. Then, such markers are used to sort hypothetic SSPCs from the pool of the isolated cells. Different assays are then employed to test the putative SSPCs self-renewal, clonogenic, and differentiation abilities. (B) Recent technological advancements, such as single-cell RNA sequencing (scRNA-seq), allows for an unbiased and ideal workflow that starts from the evaluation of the transcriptional profile and the identification of appropriate SSPCs markers right after cell isolation. After sorting them with the identified markers, SSPCs can be tested for their stem cell properties. (Created with BioRender.com ).