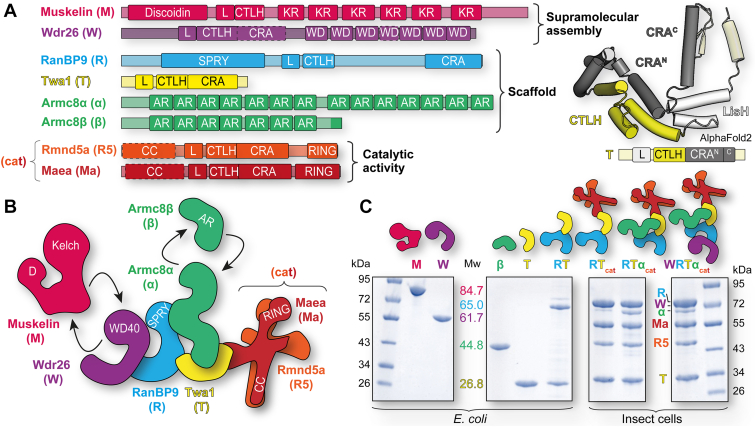
Figure 1.
Modular reconstitution of CTLH complexes.A, domain architecture of CTLH subunits based on the InterPro database and additional assignments based on AlphaFold2 predictions (dotted lines). Folding of the recurring LisH (L), CTLH, and CRA domain sequence is illustrated by the Twa1 AlphaFold2 model. B, model of the CTLH complex as defined earlier (6). Unique domains are highlighted, and arrows represent alternative subunits. C, SD-PAGE analyses of purified individual proteins and different CTLH subcomplexes after expression in either Escherichia coli or insect cells. Theoretical molecular weights (colored numerals with units in kilodalton) of the monomeric proteins as reference for the SEC–MALS analyses are indicated. AR, armadillo repeat; CC, coiled-coil region; CRA, CT-11–RanBPM; CTLH, C-terminal to lissencephaly-1 homology motif; D, discoidin; KR, kelch repeat; L, lissencephaly-1 homology; RING, really interesting new gene; SPRY, SPla and the RYanodine receptor; SEC-MALS, multiangle light scattering coupled to analytical size-exclusion chromatography; WD, WD40 repeat.