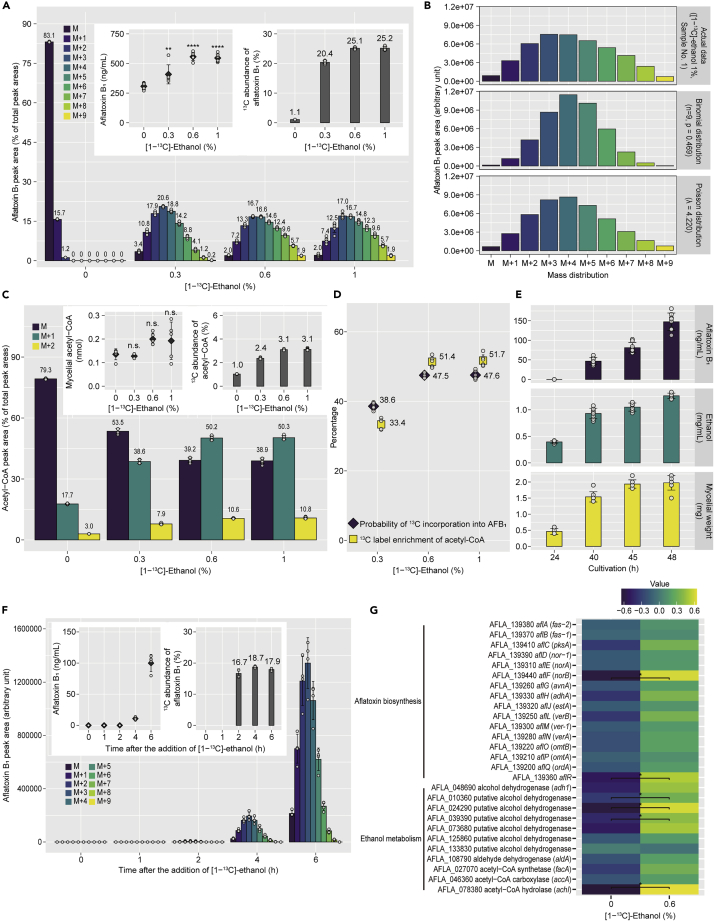
Figure 3.
Added [1-13C]-ethanol was utilized for acetyl-CoA and aflatoxin B1 biosynthesis at high rates
(A) Percentages of peak areas of aflatoxin B1 produced by A. flavus cultured with [1-13C]-ethanol; aflatoxin B1 concentrations and 13C abundance are shown in the inset panels. Mean ± SD, n = 6. ∗∗p< 0.01, ∗∗∗∗p< 0.0001 versus control, ANOVA followed by Dunnett’s test.
(B) Mass distributions generated by binomial and Poisson distribution models with estimated parameters; upper, actual data from 1% [1-13C]-ethanol–treated sample #1; middle, peak areas calculated by binomial distribution with estimated p and n = 9; lower, peak areas from [M+H]+ to [M+H+9]+ calculated by Poisson distribution with estimated λ.
(C) Percentages of peak areas of acetyl-CoA extracted from A. flavus cultured with [1-13C]-ethanol; acetyl-CoA quantities and 13C abundance are shown in the inset panels. Mean ± SD, n = 6. n.s., not significant, ANOVA followed by Dunnett’s test.
(D) Probability of 13C incorporation into aflatoxin B1 and 13C label enrichment of acetyl-CoA, estimated by binomial regression and expressed as mole percent excess (MPE),21 respectively. Mean ± SD, n = 6.
(E) Time-courses of A. flavus aflatoxin B1 production, ethanol production, and mycelial growth. Mean ± SD, n = 8.
(F) Peak areas of aflatoxin B1 collected after the addition of 0.6% [1-13C]-ethanol at 24 h cultivation; aflatoxin B1 concentrations and 13C abundance are shown in the inset panels. Mean ± SD, n = 4.
(G) Heatmap of relative gene expression patterns, determined by RT-qPCR 6 h after the addition of ethanol at 24 h cultivation. mRNA levels were standardized, and averages of n = 8 are shown. ∗p< 0.05, unpaired t test followed by the two-stage step-up procedure of Benjamini, Krieger, and Yeku to control the false discovery rate at 0.1.
See also Figure S3.