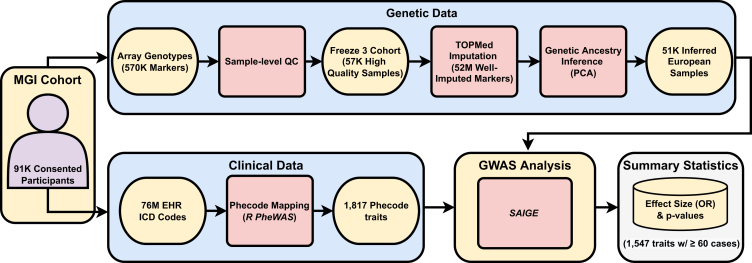
Figure 1.
Overview of the Michigan Genomics Initiative (MGI) resource and analysis
MGI currently consists of ∼91,000 participants recruited while seeking care at the Michigan Medicine health system. Recruitment is predominantly through the Department of Anesthesiology during inpatient surgical encounters. Participants agree to link a blood sample obtained during consent with their electronic health records for broad research purposes. Genotypes for ∼570,000 genetic variants are obtained from DNA extracted from the blood sample using a customized Illumina Infinium CoreExome-24 array. In this article, we describe the MGI “Freeze 3” cohort consisting of ∼57,000 samples having passed sample-level quality control filtering and imputed for >50 million variants using the TOPMed reference panel. We extracted all available International Classification of Disease (ICD) diagnosis codes from patient electronic health records and mapped to broader dichotomous phecode traits using the PheWAS software. We performed GWASs within a subset of ∼51,000 European-inferred samples from the Freeze 3 cohort using a linear mixed-effect regression model implemented in the SAIGE software. We report results and share GWAS summary statistics for 1,547 traits with ≥60 cases.