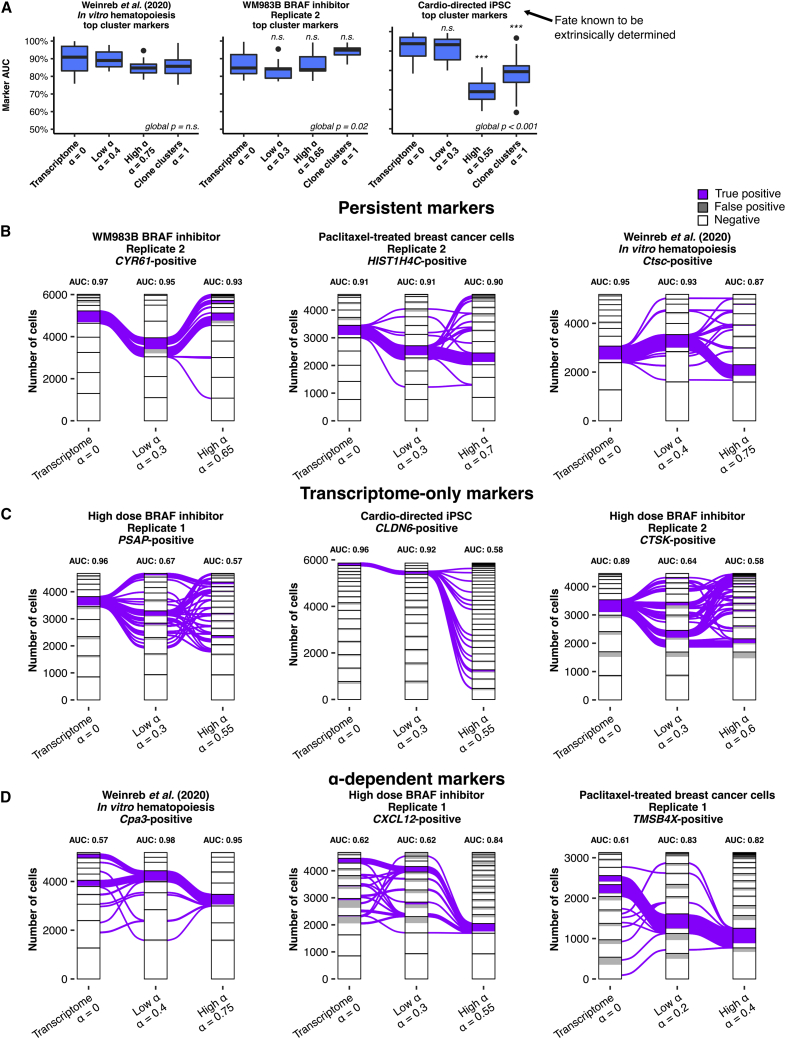
Figure 2.
Manipulation of α reveals turnover of cluster markers
(A) AUC values for the top marker per cluster at zero, low, high, and maximum (α = 1) values of α for multiple samples, an in vitro murine hematopoiesis assay at day 2, a BRAF inhibitor-treated clonal melanoma cell line WM989B, and directed differentiation of induced pluripotent stem cells (iPSCs) toward a cardiomyocyte fate, a system where extrinsic determinants of cell fate are expected be dominant over clone barcodes.18 “Global p” indicates the p value for the non-parametric Kruskal-Wallis test with Bonferroni correction. When “global p” was less than 0.05, pairwise Wilcox tests were performed with Bonferroni correction. Annotations above boxes indicate the corrected Wilcox test p value compared with α = 0 (n.s., not significant; ∗∗∗p < 0.001).
(B–D) Sankey diagram including all barcoded cells from various samples (see STAR Methods) depicting transcriptome clusters, low α clusters, and high α clusters. Cells positive for the respective marker that are present in the cluster of interest are marked purple (“True Positive”) while cells positive for the marker but not present in the cluster of interest are marked gray (“False Positive”). Positivity thresholds were determined as described in STAR Methods. Area-under-the-curve (AUC) for the marker is annotated above the cluster nodes. Representative markers are chosen to demonstrate markers whose classifier strength persists across α values (B), markers that are only strong in transcriptome clusters (C), and markers that are stronger at high and low α values (D).