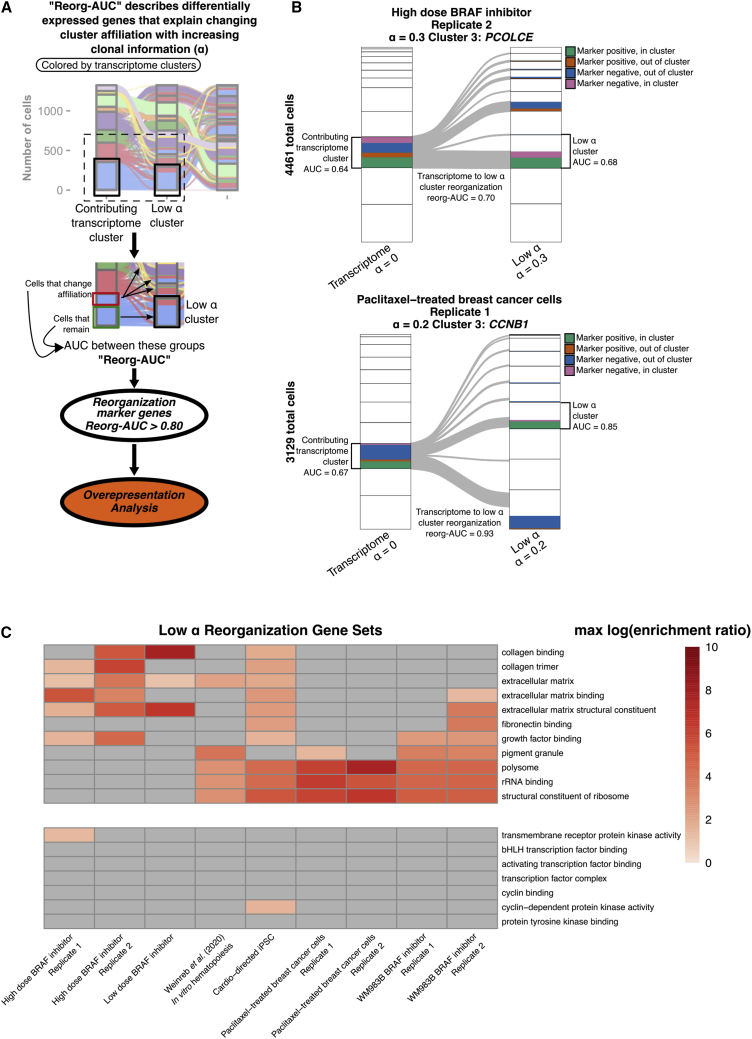
Figure 3.
Reorganization markers are enriched in translation and extracellular matrix-associated gene sets
(A) Schematic depicting approach to differential expression that explains cluster rearrangements with α modulation. For each hybrid α cluster and contributing transcriptome cluster pair, differential expression analysis is performed between cells inside and outside the α cluster, yielding an AUC for reorganization (reorg-AUC). Geneset overrepresentation analysis was then performed on these groups of differentially expressed genes on reorganizing cells with reorg-AUC >0.80 (see STAR Methods).
(B) Representative Sankey diagrams showing a low α cluster and a contributing transcriptome cluster with colors indicating proportion of cells classified by the marker and the associated cluster AUCs and reorg-AUC.
(C) Heatmap demonstrating maximum log enrichment ratio for gene sets significantly enriched by overrepresentation analysis of reorganization markers across nine different data sources. All gene sets enriched in three or more samples are shown (top) as well as several commonly explored gene sets as negative controls (bottom). Gray tiles indicate no statistically significant enrichment in the sample (false discovery rate >0.05).