Figure 2.
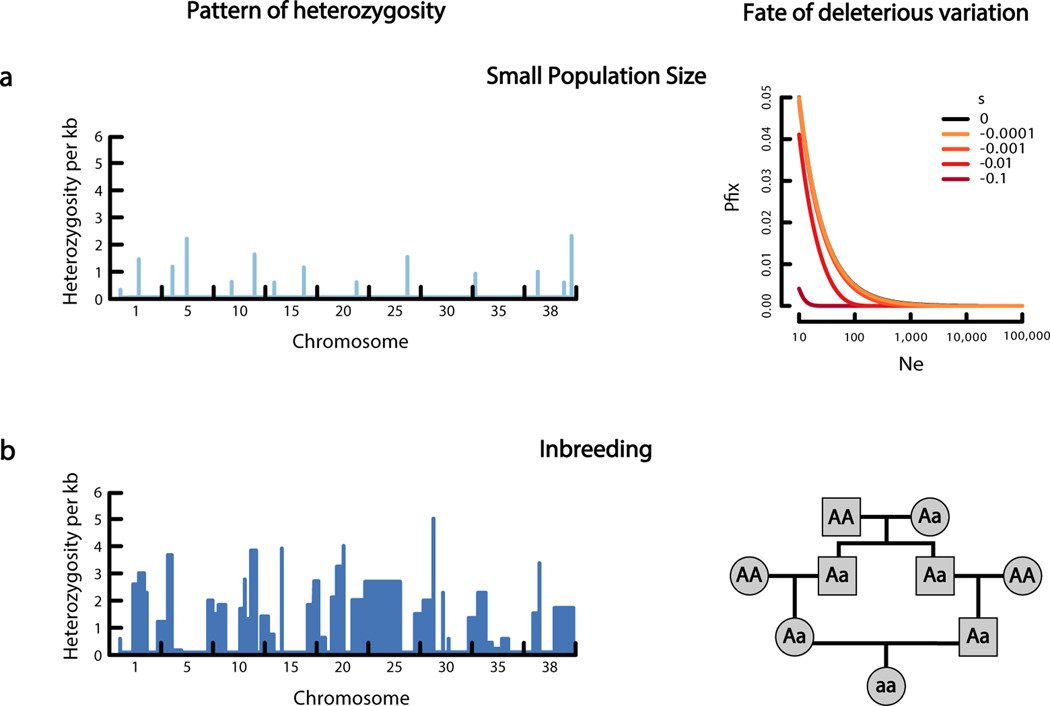
Genetic drift and inbreeding have different effects on the genome and on deleterious variation. (a, left) Genetic drift associated with a small population size leads to low heterozygosity across the genome. (right) Genetic drift also allows weakly deleterious mutations to become fixed in the population, contributing to drift load. The right-hand panel shows the probability of fixation for a new mutation with s specified by the different lines in a population of the size denoted on the x-axis. Probabilities were calculated using equation 10 in Reference 128. (b, left) Inbreeding can lead to a sawtooth pattern of heterozygosity across the genome. Regions of low heterozygosity correspond to the regions of the genome that likely are copies of the same ancestral chromosome inherited from both parents. (right) Inbreeding results in an increase in the probability of an ancestral chromosome (denoted by a) being inherited from both the maternal and paternal lineage, increasing homozygosity. Thus, recessive deleterious mutations have a higher probability of becoming homozygous and thereby affecting fitness.