ABSTRACT
Chlamydia trachomatis is the most common cause of infectious blindness and sexually transmitted bacterial infection globally. C. trachomatis contains a conserved chlamydial plasmid with eight coding sequences. Plasmid-cured Chlamydia strains are attenuated and display reduced infectivity in cell culture and in vivo genital infection of female mice. Mutants that do not express the plasmid-encoded proteins Pgp3, a secreted protein with unknown function, or Pgp4, a putative regulator of pgp3 and other chromosomal loci, display an infectivity defect similar to plasmid-deficient strains. Our objective was to determine the combined and individual contributions of Pgp3 and Pgp4 to this phenotype. Deletion of pgp3 and pgp4 resulted in an infectivity defect detected by competition assay in cell culture and in mice. The pgp3 locus was placed under the control of an anhydrotetracycline-inducible promoter to examine the individual contributions of Pgp3 and Pgp4 to infectivity. Expression of pgp3 was induced 100- to 1,000-fold after anhydrotetracycline administration, regardless of the presence or absence of pgp4. However, secreted Pgp3 was not detected when pgp4 was deleted, confirming a role for Pgp4 in Pgp3 secretion. We discovered that expression of pgp3 or pgp4 alone was insufficient to restore normal infectivity, which required expression of both Pgp3 and Pgp4. These results suggest Pgp3 and Pgp4 are both required for infectivity during C. trachomatis infection. Future studies are required to determine the mechanism by which Pgp3 and Pgp4 influence chlamydial infectivity as well as the potential roles of Pgp4-regulated loci.
KEYWORDS: Chlamydia, intracellular bacteria, pathogenesis, virulence factors
INTRODUCTION
Chlamydia trachomatis is the leading cause of infectious blindness globally (1) and the most common bacterial sexually transmitted infection in the United States and around the world. In 2020, C. trachomatis caused almost 1.6 million urogenital infections in the United States, and the number of infections has continued to trend upward over the last 10 years (2). In women, C. trachomatis can ascend to infect the upper genital tract, which can lead to long-term sequelae such as pelvic inflammatory disease, ectopic pregnancy, and/or infertility.
Chlamydiae are obligate intracellular bacteria with a biphasic developmental cycle (3). To begin the cycle, a nonreplicative elementary body (EB) attaches to the host cell surface and induces bacterial uptake by secreting chlamydial effectors into the host cell cytosol. Once inside, the EB transitions to a reticulate body (RB) and begins to replicate inside a membrane-bound vacuole called the inclusion. Throughout the developmental cycle, chlamydiae continue to secrete effectors into the inclusion and across its membrane into the host cell cytosol to modulate host cell processes and create an environment conducive to bacterial survival and replication. Ultimately, RBs transition to EBs and the bacteria are released by cell lysis or inclusion extrusion (4).
The conserved 7.5-kb chlamydial plasmid plays a vital role in Chlamydia infection and disease. Loss of the chlamydial plasmid is pleiotropic. Plasmid-deficient chlamydiae display an infectivity defect in cell culture and in a mouse model of genital tract infection (5–9), do not accumulate glycogen in their inclusions (5, 10), fail to activate Toll-like receptor 2 (6, 9), and elicit lower levels of cytokines from infected cells (6, 9). Plasmid-deficient Chlamydia muridarum caused significantly less pathology in a mouse genital tract infection model (6, 8), while plasmid-deficient C. trachomatis was rapidly cleared from the murine genital tract model after transcervical infection (11) and exhibited accelerated clearance and caused less pathology in a nonhuman primate trachoma model (12).
The development of techniques for genetically manipulating and transforming chlamydiae has paved the way for studies examining the roles of each of the eight open reading frames (pgp1 to pgp8) encoded on the chlamydial plasmid (13, 14). pgp1, -2, -6, and -8 are essential for plasmid maintenance (15). The proteins encoded by pgp5 and pgp4 were identified as potential transcriptional regulators. Pgp5 is a negative regulator that contributes to hydrosalpinx formation in mice (16, 17). Pgp4 is important for expression and secretion of multiple chromosomal loci as well as plasmid-encoded pgp3 (15, 18). The pgp3 locus encodes a trimeric protein that is secreted into the host cell cytosol late in infection (19). Deletion of either pgp3 or pgp4 resulted in decreased bacterial burden in the lower and upper genital tract of infected female mice (20, 21). While these studies suggest both Pgp3 and Pgp4 are involved in plasmid-associated infectivity, it has been assumed that pgp4-deficient Chlamydia is less infectious than wild-type because it does not express Pgp3. However, the functions of most Pgp4-regulated loci are unknown, and their potential contribution to chlamydial infectivity has never been assessed.
The objective of this study was to explore possible roles for proteins encoded by Pgp4-regulated chromosomal loci in infectivity either independently or in cooperation with Pgp3. We engineered three C. trachomatis recombinants, each carrying a modified chlamydial plasmid: a pgp3- and pgp4-deficient strain and two strains expressing anhydrotetracycline (aTc)-inducible pgp3 in the presence and absence of pgp4. We investigated the impact of pgp4 deletion on Pgp3 secretion and examined the infectivity of each strain in cell culture using a competition assay. Our findings confirmed a role for Pgp4 in Pgp3 secretion and revealed that Pgp3 or Pgp4 alone are insufficient to mediate plasmid-associated infectivity. Based upon these results and previous studies, we hypothesize a novel mechanism for the role of Pgp3 secretion in chlamydial infectivity.
RESULTS
Pgp3 and Pgp4 deficiency results in defects in glycogen accumulation but does not impact growth.
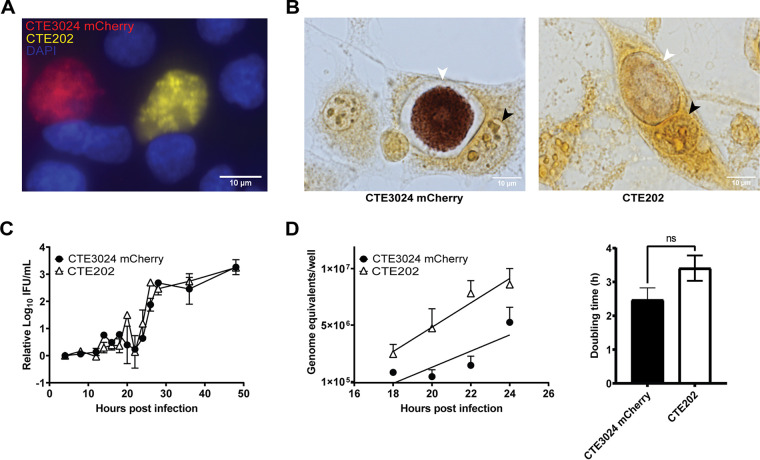
Pgp3 and Pgp4 have been implicated as mediators of multiple plasmid-associated phenotypes, including loss of glycogen accumulation in C. trachomatis (via Pgp4) (22) and infectivity (via Pgp3 and Pgp4) (20, 21). To examine the combined impact of pgp3 and pgp4 deletion on these plasmid-associated phenotypes, we first deleted pgp3 and pgp4 from a chlamydial shuttle vector expressing green fluorescent protein (GFP). This plasmid was then transformed into CTE3024, a C. trachomatis serovar E strain isolated from a woman with confirmed endometrial infection. The resulting pgp3- and pgp4-deficient and GFP-positive strain was named CTE202. We observed no difference in inclusion morphology between CTE3024 mCherry (wild type with respect to plasmid loci) and CTE202 (Fig. 1A), but CTE202 did not accumulate glycogen in its inclusion (Fig. 1B). Cells infected at an multiplicity of infection (MOI) of 0.7 were harvested at designated times throughout the chlamydial developmental cycle. Examination of growth by the ability to form infectious progeny (Fig. 1C) revealed no significant difference between CTE3024 mCherry and CTE202. Both strains displayed a similar “eclipse” phase of approximately 22 h and produced similar numbers of infectious progeny starting around 22 h. We estimated the genome replication rate in reticulate bodies between 18 h and 24 h postinfection (p.i.) as a measure of bacterial division, reasoning that reticulate body replication would be highest and most consistent in the window of development immediately prior to EB generation, while limiting confounding by nonreplicating EB. The genome replication rate was similar between CTE3024 mCherry and CTE202 (Fig. 1D), and we did not detect a significant difference in doubling times between the two strains (mean doubling time ± standard deviation [SD] of CTE3024 mCherry, 2.5 ± 0.7 h, versus CTE202, 3.4 ± 0.7 h). These results showed deletion of pgp3 and pgp4 did not impact chlamydial growth kinetics for this human clinical isolate.
FIG 1.
CTE202 does not accumulate glycogen and grows similar to wild type in vitro. (A) L929 cells infected with CTE3024 mCherry (red) or CTE202 (yellow) at an MOI of 0.5 showed no difference in inclusion morphology at 40 h p.i. Nuclei (blue) were visualized using NucBlue live cell stain, and cells were live imaged with a 40× objective using the EVOS M7000 microscope. (B) Despite no visible difference in inclusion morphology, CTE3024 mCherry (left) inclusions stained positively for glycogen by iodine staining 40 h p.i., while CTE202 (right) inclusions did not. Black arrows indicate nuclei, and white arrows indicate chlamydial inclusions. Infected cells were imaged at 1,000×. Consistent with inclusion morphology, there was no difference in growth between CTE3024 mCherry or CTE202. Growth curves were plotted from L929 cells infected with CTE3024 mCherry or CTE202. (C and D) Samples were taken at designated time points, and infectious progeny quantified as inclusion forming units (IFU) per milliliter (mL) (C) or genome equivalents (D). IFU data were normalized to 4-h samples prior to calculating the average. Growth rate is represented as the slope of the linear regression during exponential growth (18 to 24 h p.i.). Data points represent means ± standard deviations (SD) of two replicates assayed in duplicate. P = 0.33 by two-way ANOVA with Šídák’s multiple comparisons (IFU data), and P = 0.38 by linear regression (genome equivalents data). Doubling time was calculated from the genome equivalents data at the 18-h and 24-h time points. Graphs show means ± SD from four replicates. ns, not significant; P = 0.12 by unpaired t test. Cells shown in all four panels were infected using centrifugation.
Loss of pgp3 and pgp4 recapitulates the infectivity defect associated with plasmid deficiency in cell culture and a mouse model of infection.
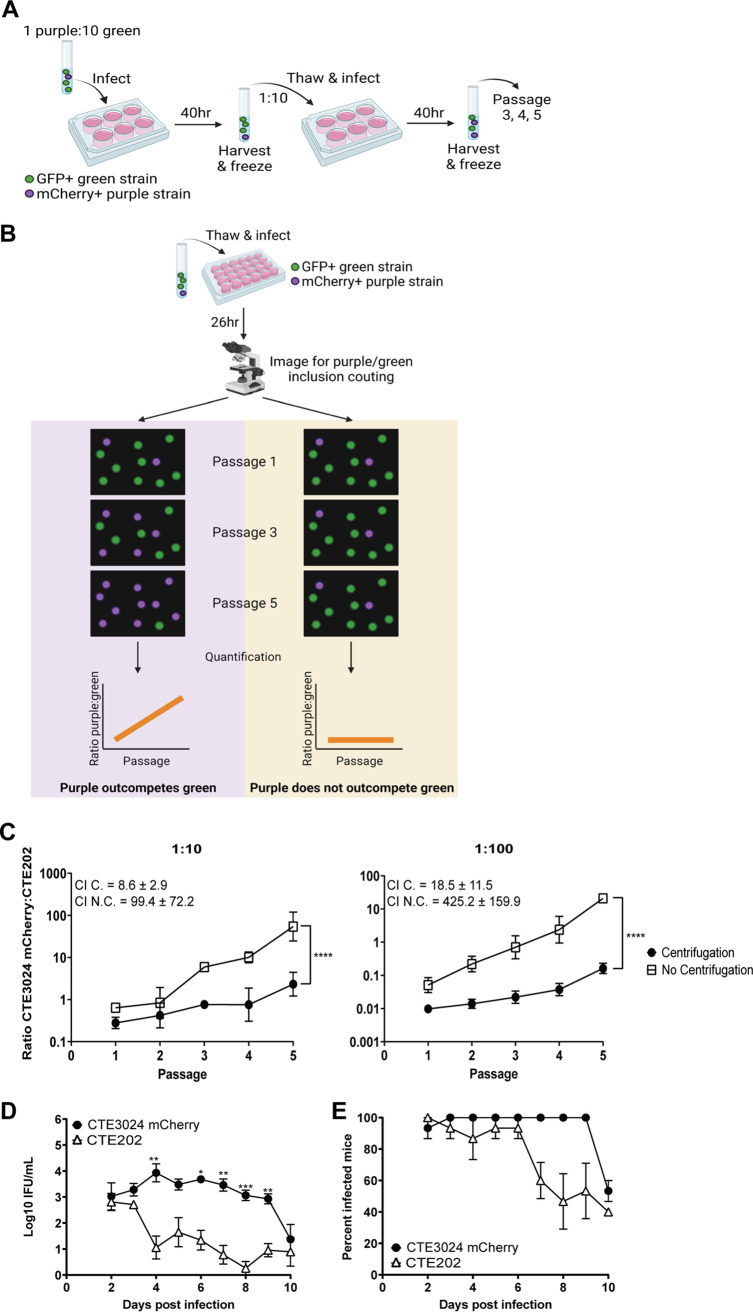
The parent strain CTE3024 and its derivatives do not form readily observable plaques in cell culture, preventing use of plaque-based methods (5, 10) to assess infectivity. Russell et al. suggested that in vitro competition assays more accurately recapitulated the infectivity defect observed in plasmid-deficient Chlamydia muridarum when results from mouse genital tract studies and cell culture studies were compared (23). Therefore, we compared the infectivity of CTE202 to CTE3024 mCherry using an in vitro competition assay (23) (Fig. 2A). CTE3024 mCherry and CTE202 were mixed in ratios of 1:10 and 1:100, rendering CTE3024 mCherry at a disadvantage. Chlamydial mixtures were infected onto monolayers with and without centrifugation, because centrifugation can overcome the plasmid-associated infectivity defect in cell culture (5). Chlamydiae were harvested at 40 h p.i. to reduce possible confounding effects from differential cell lysis (24) and frozen for ease of experimentation. A portion of each passage was infected onto a new monolayer to continue the culture with synchronous infection. The mCherry-expressing CTE3024 mCherry inclusions and GFP-expressing CTE202 inclusions were quantified using live imaging following each passage (Fig. 2B). Additional details can be found in Materials and Methods. The ratio of CTE3024 mCherry to CTE202 increased significantly over the course of five passages (Fig. 2C) and resulted in a competitive index (CI) ranging between 8.6 ± 2.9 and 18.5 ± 11.5, suggesting CTE3024 mCherry retains a slight advantage over CTE202 at baseline. However, without centrifugation, CTE3024 mCherry became the predominant strain in the culture by passage 3 (1:10) or 4 (1:100), with a 10-fold-higher CI. The slope of this line significantly differed from 0, reflecting asymmetric expansion of the CTE3024 mCherry population with multiple passages, and was significantly steeper than that observed with centrifugation, suggesting the advantage held by CTE3024 mCherry was much greater in the absence of centrifugation.
FIG 2.
CTE202 displays an infectivity defect in cell culture and in a mouse model of genital tract infection. (A) Cartoon depicting the workflow used for competition assays. GFP+ (depicted in green) and mCherry+ (depicted in purple) strains were mixed together and infected onto cell monolayers. At 40 h p.i., the monolayers were harvested and passaged onto new cell monolayers. This process continued for a total of five passages. (B) Schematic depicting the imaging and quantitation procedure used for competition assays. A portion of each passage from panel A was infected onto cell monolayers. GFP+ inclusions and mCherry+ inclusions were imaged and quantified, and the ratio of GFP+:mCherry+ was calculated. Graphing of ratios resulted in a line with a positive slope different from 0 (evidence of competition) or a flat line (no competition). The images in panels A and B were created using BioRender. (C) Quantification of the ratio of CTE3024 mCherry to CTE202 from passages of competition assays showed CTE3024 mCherry outcompeted CTE202 in the absence of centrifugation, but the ability to outcompete was significantly reduced by centrifugation. Data points represent the average ratios from two technical replicates ± SD. Slopes of the lines were compared to 0 (P ≤ 0.0001 [1:10, 1:100]) or to each other (P ≤ 0.0001 [1:10, 1:100]) by simple linear regression. Graphs shown are representative of 3 experiments. C., centrifugation; N.C., no centrifugation. (D) Similar to cell culture, CTE3024 mCherry was more infectious than CTE202 in a mouse model of infection. Five mice per group were infected transcervically with 5 × 106 IFU/mL CTE3024 mCherry or CTE202. Infection was monitored via cervico-vaginal swab and quantified as IFU. Data points represent the mean ± standard error of the mean (SEM) of all mice from one of three representative experiments. P < 0.0001 by two-way ANOVA with repeated measures and Šídák’s multiple comparisons. *, P < 0.05; **, P < 0.005; ***, P < 0.0005. (E) Points represent the mean percent infected mice ± SD of three independent experiments. P = 0.004 by Wilcoxon signed-rank test.
In vitro competition assays mimic the serial nature of in vivo infection but lack the diverse cell types and stressors encountered by chlamydiae during human or animal infection. C. trachomatis genital infection of mice results in an acute, self-resolving infection. Comparisons of intravaginal and transcervical inoculation of mice with C. trachomatis serovar D showed transcervical infection resulted in higher bacterial burdens during the early days of infection (25). Therefore, to determine if CTE202 also exhibited an infectivity defect in vivo, we utilized the C. trachomatis mouse model of transcervical infection (25). Bacterial burdens in mice infected with CTE3024 mCherry remained consistent from days 2 to 9 before decreasing sharply at day 10 (Fig. 2D). Despite similar bacterial burdens in CTE3024 mCherry-infected mice and CTE202-infected mice on days 2 to 3 p.i., CTE202 burden fell on day 4 and bacterial loads were significantly lower for CTE202 compared to CTE3024 mCherry on days 4 to 9. CTE202 also appeared to be more rapidly cleared because the average percentage of infected mice over the first 10 days from three independent experiments was overall significantly lower in mice infected with CTE202 than in those infected with CTE3024 mCherry (Fig. 2E). Together, these results suggested CTE202 exhibits an infectivity defect in cell culture and in vivo and supports a role for pgp3 and/or pgp4 in C. trachomatis infectivity.
An anhydrotetracycline-inducible system can be used to isolate pgp3 expression from the Pgp4-dependent regulon.
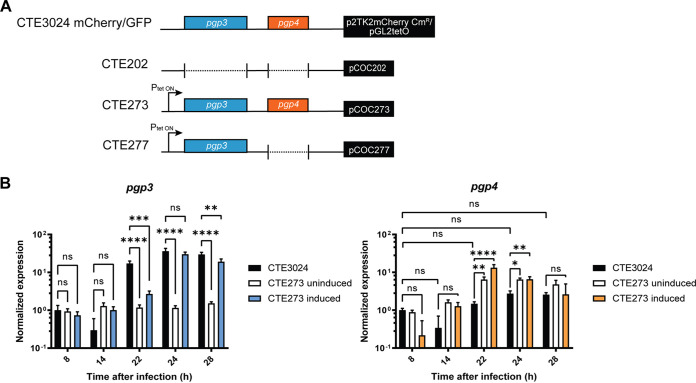
Pgp3 and Pgp4 have been implicated in plasmid-associated infectivity by deletion of both loci (this study) and single deletion of each gene (20, 21). However, Pgp4 is involved in pgp3 expression (15), impeding examination of the independent contributions of pgp3 and pgp4 to infectivity. To overcome this barrier, we rendered pgp3 independent of pgp4 by placing it under the control of an aTc-inducible promoter so that we could determine the consequences of expression in the presence and absence of pgp4 (Fig. 3A). We observed that pgp3 expression increased 10- to 100-fold above baseline within 30 min of adding 25 ng/mL aTc for CTE273 and CTE277 (Fig. S3A). The level of pgp3 expression decreased slightly after 30 min but remained steady overall from 2 to 6 h postinduction. Exposure to 5 ng/mL aTc induced pgp3 expression to lower, but still detectable, levels in CTE273 at 30 min postinduction and in CTE277 at 30 min, 4 h, and 6 h postinduction. Induction of protein expression with aTc can have deleterious effects on chlamydial growth (26, 27). We observed a significant reduction in the yield of infectious progeny following exposure to 25 ng/mL aTc from the recombinants we had constructed, but not following exposure to 5 ng/mL aTc (Fig. S3B). We also compared induced pgp3 transcription levels to native pgp3 expression. Transcription data were normalized to the 8-h time point collected from CTE3024 mCherry, because Pgp3 is not detected at this time point in wild-type C. trachomatis (18, 19). Expression of pgp3 was undetectable in CTE3024 mCherry at 14 h p.i. but increased at 22 h and remained stable through 28 h p.i. (Fig. 3B). In the absence of aTc, pgp3 expression in CTE273 did not increase above baseline levels at any time point measured. After administration of aTc at 22 h p.i., we detected elevated pgp3 transcripts at 24 and 28 h to levels similar to those detected in CTE3024 mCherry, indicating that induction of pgp3 using 5 ng/mL was comparable to pgp3 induction from the native promoter. Finally, we assessed transcription of pgp4 to determine if aTc induction of pgp3 caused transcriptional readthrough to alter pgp4 expression, as only ~70 bp separate the end of pgp3 from the transcriptional start of pgp4. Overall, expression of pgp4 in CTE3024 mCherry and CTE273 was constitutive. Nevertheless, the mutant expressed slightly higher levels of pgp4 than CTE3024 mCherry. We observed a significant increase in pgp4 expression at 22 and 24 h p.i. in CTE273, compared to CTE3024 mCherry, but this increase was observed in CTE273 with and without aTc. Further, the levels of pgp4 at these time points did not significantly differ from basal levels detected at 8 h p.i. This suggested induction of pgp3 expression did not lead to artifactual pgp4 expression.
FIG 3.
Induction of pgp3 with anhydrotetracycline does not influence expression pgp4. (A) Close-up of pgp3 and pgp4 loci on shuttle vectors used in this study. Colored boxes represent presence of pgp3 (blue) or pgp4 (orange). Arrow designates the direction of the tet-inducible promoter upstream of pgp3. Plasmids are labeled in the black box on the right, which symbolizes the remaining plasmid DNA. Plasmids were transformed into CTE3024 to create CTE202, CTE273, and CTE277, as shown by the strain name labels on the left. (B) Transcription of pgp3 and pgp4 in CTE3024 mCherry and CTE273 was measured following induction with 5 ng/mL of aTc at 22 h p.i. Transcription was quantified by quantitative reverse transcription-PCR. Graphs show the average normalized expression of two biological replicates assayed in triplicate. Gene expression was normalized to that of 23S and the CTE3024 mCherry 4-h sample. ns, not significant, P > 0.05; *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.001 by two-way ANOVA with Tukey’s multiple comparisons.
Pgp4 is required for Pgp3 secretion to the host cell cytosol during C. trachomatis infection.
Pgp3 is a secreted effector that can be detected as puncta localized to the chlamydial inclusion as well as in the host cell cytosol during infection (19). Immunofluorescent staining of HeLa cells infected with CTE3024 mCherry showed bright punctate staining of Pgp3 localized to the inclusion as well as diffuse cytosolic staining (Fig. 4A). As expected, there was no detectable Pgp3 staining in CTE202-infected cells or in CTE277- or CTE273-infected cells grown in the absence of inducer. Induction of Pgp3 expression in CTE273 was detected as Pgp3 staining at the inclusion and in the cytosol, similar to CTE3024 mCherry. However, staining of CTE277-infected cells after induction revealed a pattern of intense Pgp3 staining localized primarily to the inclusion. Cytosolic staining of Pgp3 was only detected in approximately 15% of infected cells, versus approximately 60% in CTE3024 mCherry- and CTE273-infected cells treated with aTc (Fig. 4B). Cytosolic staining of Pgp3 was not detected in uninduced CTE273- or CTE277-infected cells. The absence of Pgp4 expression in CTE277 was confirmed by the observed lack of glycogen accumulation (Fig. S4). As expected, CTE273 inclusions accumulated glycogen similar to CTE3024 mCherry. These data confirmed a role for Pgp4 in the secretion of Pgp3 during C. trachomatis infection.
FIG 4.
Pgp4 is required for efficient secretion of Pgp3. (A) Immunofluorescent staining of HeLa cells infected with CTE3024 mCherry, CTE202, CTE273, or CTE277 showed cytosolic Pgp3 staining in CTE3024 mCherry- and CTE273-infected cells, but not CTE277-infected cells following induction with 5 ng/mL aTc at 22 h p.i. Cells were fixed at 32 h p.i., stained for Pgp3 (cyan), MOMP (red), and nuclei (blue), and imaged with a 100× objective on the EVOS M7000 microscope. Insets show closeups of cytosolic Pgp3 staining present in CTE3024 mCherry and CTE273, but not CTE202 or CTE277. (B) The number of infected cells containing Pgp3 in the host cell cytosol was quantified from induced and uninduced CTE3024 mCherry-infected HeLa cells and induced CTE273- and CTE277-infected HeLa cells. A total of 100 to 150 infected cells from at least three microscopic fields were quantified from each replicate. Graph shows the average of two replicates ± SD and is representative of three experiments. ns, not significant, P > 0.05; **, P < 0.005 by one-way ANOVA with Tukey’s multiple comparisons.
Pgp3 and Pgp4 are required for infectivity during C. trachomatis infection.
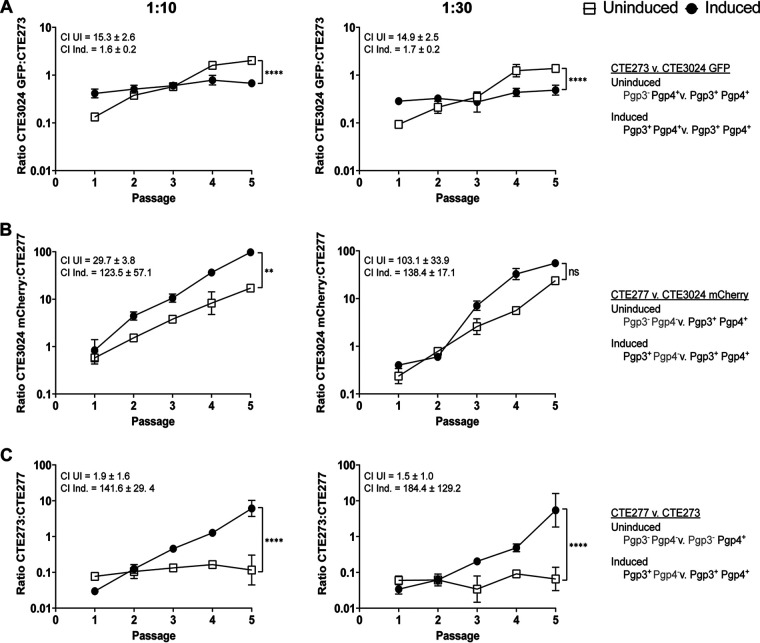
Following optimization of aTc-mediated induction of Pgp3 expression, we examined the roles of Pgp3 and Pgp4 in plasmid-associated infectivity using the cell culture-based competition assay. Competing CTE273 and CTE3024 GFP under noninducing conditions showed expansion of CTE3024 GFP (Fig. 5A), yielding CI values of 15.3 ± 2.6 (1:10) and 14.9 ± 2.5 (1:30). These CIs were similar to those obtained for CTE3024 mCherry in competition with CTE202 (Fig. 2C), again suggesting that a lack of Pgp3 contributes to an infectivity defect in this assay. In contrast, aTc induction of Pgp3 in CTE273 greatly reduced the advantage held by CTE3024 GFP under uninduced conditions (CIs,1.6 and 1.7). When the pgp4-deficient CTE277 was placed in competition against CTE3024 mCherry, we observed that CTE3024 mCherry displayed a significant advantage under inducing and noninducing conditions over five passages, with CI values ranging from 29.7 to 138.4, depending upon the condition tested (Fig. 5B). CTE3024 mCherry outgrowth occurred significantly faster under induced conditions at a starting ratio of 1:10. However, this result was not observed when a ratio of 1:30 was used, suggesting that expression of Pgp3 without Pgp4 does not restore infectivity. Finally, we placed CTE273 and CTE277 in competition and observed no asymmetric expansion of either strain under noninducing conditions (Fig. 5C). In contrast, when pgp3 expression was induced, CI values increased from 1.5 to 1.9 under noninducing conditions to 141.6 to 184.4 with induction and resulted in a significantly steeper slope than that generated under noninducing conditions. Taken together, these data show that Pgp3 alone is insufficient to mediate plasmid-associated infectivity and Pgp4 is required for plasmid-associated infectivity in C. trachomatis, independent of pgp3 transcriptional regulation.
FIG 5.
Pgp3 and Pgp4 are required for plasmid-associated infectivity. Competition assays with starting ratios of 1:10 (left) or 1:30 (right) for CTE273:CTE3024 GFP (A), CTE277:CTE3024 mCherry (B), or CTE277:CTE273 (C) showed strains expressing Pgp3 and Pgp4 outcompeted strains expressing Pgp3 or Pgp4 alone. GFP-expressing and mCherry-expressing inclusions were quantified following each passage, and ratios were calculated. Data points represent mean ratios ± SD from two replicates. Competitive indices are means ± SD. Graphs are representative of two experiments. Slopes of the lines were compared to 0: P < 0.0001 (1:10 uninduced, 1:30 uninduced), P = 0.007 (1:10 induced), P = 0.027 (1:30 induced) (A); P < 0.0001 (1:10 uninduced, 1:10 induced, 1:30 uninduced, and 1:30 induced) (B); P = 0.31 (1:10 uninduced), P < 0.0001 (1:10 induced, 1:30 induced) P = 0.6873 (1:30 uninduced) (C); or to each other: ****, P < 0.0001 (1:10, 1:30) (A); **, P = 0.0007 (1:10), ns, P = 0.22 (1:30) (B); ****, P < 0.0001 (1:10, 1:30) (C); using simple linear regression. ns, not significant; UI, uninduced; Ind., induced.
OmcB and major outer membrane protein are normally expressed in plasmid-deficient C. muridarum.
Our observations that Pgp3 expression in the absence of Pgp4 did not restore infectivity and that Pgp4 was required for Pgp3 secretion suggested that secretion of Pgp3 may be required for normal infectivity. However, pgp3 transcription, and particularly Pgp3 secretion, occurs late in the developmental cycle, inconsistent with chlamydial entry and secretion of other chlamydial effectors with direct roles in infectivity (28–34). We speculated that disrupting a highly coordinated secretion mechanism coincident with the RB-EB transition could result in protein mislocalization and yield EBs with compromised infectivity. A recent study proposed that Pgp3 and other plasmid-regulated chromosomal loci are secreted via outer membrane vesicles (OMV) (18). OMV production is heavily reliant on outer membrane dynamics. We hypothesized that disruption of OMV production in strains that do not express Pgp3 and/or Pgp4 could impede membrane events important for infectious EB production, such as placement or posttranslational modification of an adhesin molecule in the outer membrane.
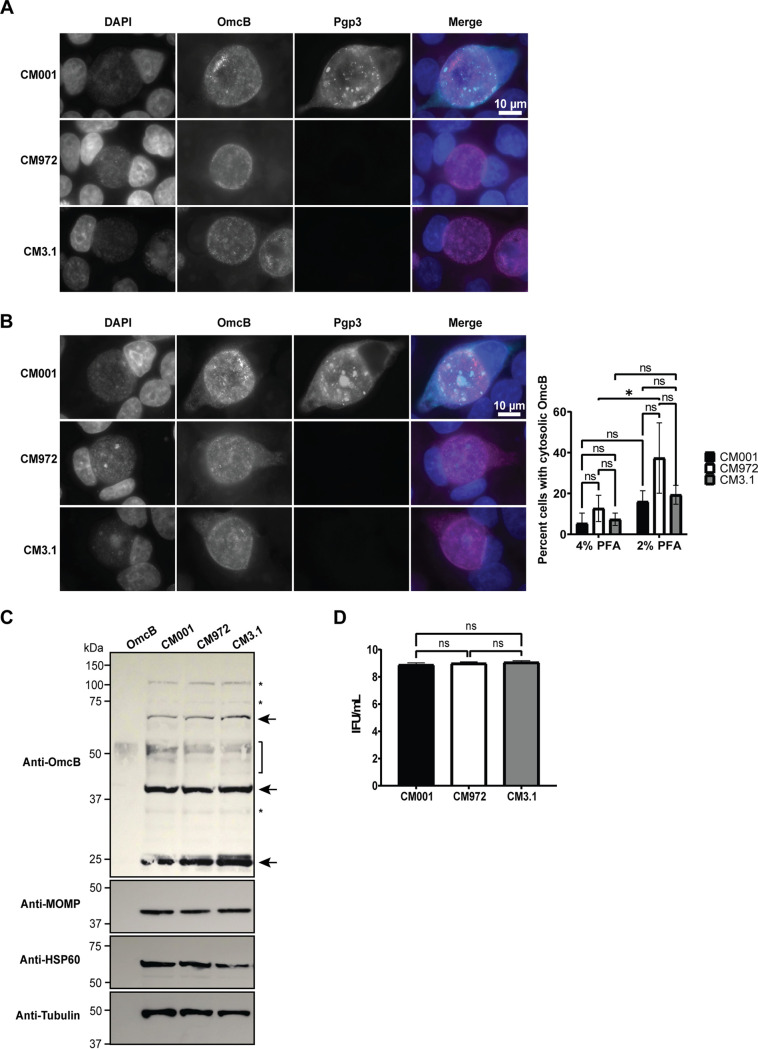
OmcB and the major outer membrane protein (MOMP) are both outer membrane proteins that have been proposed as candidate adhesins for chlamydiae (35–43). OmcB binds glycosaminoglycans on the surface of host cells (44). Published studies suggest OmcB (60 kDa) is cleaved into two fragments, a 20-kDa N-terminal fragment and a 40-kDa C-terminal fragment that is secreted into the host cell cytosol during infection (45, 46). Those authors postulated that the C-terminal fragment could be secreted via OMV (46). The significance of OmcB cleavage and subsequent secretion of the C-terminal fragment for adhesion and infection is unknown, but retention of the C-terminal fragment in the periplasm could interfere with OmcB cleavage and/or adhesin activity. MOMP is proposed to interact with epithelial cells via heparan sulfate receptors (35) and is excluded from OMV produced by wild-type chlamydiae (18). To examine OmcB and MOMP expression in plasmid-deficient chlamydiae, we utilized wild-type C. muridarum strain CM001 and two plasmid-deficient C. muridarum strains, CM972 and CM3.1. CM972 exhibits an infectivity defect in cell culture (5) and in vivo (6). However, CM3.1 contains a suppressor mutation (23) which restores wild-type infectivity (6, 23). We expected that any deficit detected in adhesin expression or processing by CM972 compared to CM001 would not be present in CM3.1. We reasoned that if we observed this pattern, we could attribute the differences in adhesin expression or processing between CM001 and CM972 to infectivity, rather than an unrelated consequence of plasmid deficiency. A plasmid-deficient suppressor mutant with restored infectivity has not been described for C. trachomatis, precluding our ability to observe restoration of adhesin expression or processing phenotypes in a plasmid-deficient strain with wild-type infectivity. When we examined OmcB protein expression by immunofluorescent staining, we observed OmcB staining localized to the bacteria in most infected cells (Fig. 6A), despite using the published fixation and permeabilization conditions previously used to detect secreted OmcB (46). Detection of cytosolic Pgp3 in CM001-infected cells confirmed that proteins secreted into the host cell cytosol could be detected using these staining conditions. Nevertheless, we observed low levels of diffuse OmcB staining in a subset of infected cells that appeared to be localized to the cytosol (Fig. 6B). The percentage of cells containing cytosolic OmcB staining trended higher in CM972-infected cells, compared to CM001- and CM3.1-infected cells, but was not significantly different. Cytosolic OmcB staining was reduced in CM972-infected cells when cells were fixed with 4% paraformaldehyde (PFA) at 37°C, suggesting OmcB localization to the cytosol in cells fixed slowly with 2% PFA at room temperature is possibly an artifact arising from these fixation conditions (47).
FIG 6.
OmcB expression and processing does not differ between wild-type and plasmid-deficient strains. (A) Immunofluorescent staining of L929 cells infected with CM001 (wild type), CM972 (plasmid deficient), or CM3.1 (plasmid deficient, no infectivity defect) showed no difference in OmcB localization. Cells were fixed 40 h p.i. and stained with OmcB antiserum (green) and 4′,6-diamidino-2-phenylindole (blue). Images were taken with a 100× objective on an EVOS M7000 digital microscope. (B) A proportion of infected cells from panel A showed OmcB staining in the cytosol. Quantification of 50 inclusions per group per replicate showed a trend for increased OmcB localization in CM972-infected cells in cells fixed with 2% PFA. Bars represent means ± SD of three replicates. *, P = 0.03 by two-way ANOVA with Tukey’s multiple-comparisons test. (C) CM001 and plasmid-deficient CM972 and CM3.1 expressed similar levels of OmcB and MOMP as measured by immunoblotting. A 25-μg sample of whole-cell lysate was run in each lane, and 500 ng recombinant OmcB was included as a control. Membranes were blotted with OmcB immune serum and antibodies against C. muridarum MOMP, HSP60, and tubulin. Arrows indicate OmcB, the brackets show possible degradation products in lanes containing lysates, and the asterisk designates nonspecific bands. Images were cropped to isolate relevant bands and samples from a single replicate. Uncropped images can be found in Fig. S5 in the supplemental material. The blot is representative of n = 3 experiments. (D) Quantification of EBs harvested from samples described for panel C, showing similar numbers of EBs were present in each sample. Graphs depict the means ± SD of two replicates assayed in duplicate and are representative of n = 2 experiments. ns, nonsignificant; *, P = 0.15 by one-way ANOVA with Tukey’s multiple-comparisons test.
Chlamydial protease activity factor (CPAF) cleaves OmcB in cell culture (45). Therefore, to mitigate potential confounding events arising from this activity, we treated the cells with the CPAF inhibitor lactacystin (48) prior to lysis and harvested lysates in hot 1% SDS, which inhibits OmcB cleavage by CPAF postlysis (48, 49). We examined whole-cell lysates from infected cells by immunoblotting to capture potential differences in full-length OmcB and the secreted C-terminal fragment (46). We were unable to detect differences in total OmcB protein or cleavage patterns between lysates derived from CM001-, CM972-, or CM3.1-infected cells (Fig. 6C). Furthermore, we detected no difference in the total amount of MOMP present between the strains. Enumeration of infectious progeny by IFU showed similar numbers of EBs were present in each sample tested (Fig. 6D). Together, these data demonstrate that MOMP and OmcB protein expression and the OmcB cleavage profile do not differ between plasmid-deficient and wild-type strains of C. muridarum. However, slow fixation results in increased cytosolic OmcB in CM972-infected cells, compared to faster fixation methods.
DISCUSSION
In this study, we examined the combined and individual roles of Pgp3 and Pgp4 in chlamydial infectivity. We determined that expression of Pgp3 or Pgp4 alone was insufficient to restore plasmid-associated infectivity in C. trachomatis and we have provided the first evidence that the role of Pgp4 is independent of its regulatory contribution to pgp3 expression.
Deleting pgp3 and pgp4 in C. trachomatis recapitulated the infectivity defect first detected in plasmid-deficient C. muridarum (5), providing additional evidence of a role for Pgp3 and Pgp4 in the infection of host cells. The infectivity defect was significantly reduced when cells were infected by centrifugation, an observation mostly consistent with our previous study, in which a plasmid-associated infectivity deficit was overcome by centrifugation (5). However, in the current study, wild-type CTE3024 mCherry retained a slight advantage over pgp3- and pgp4-deficient CTE202 over the course of five passages, even with centrifugation-assisted infection, whereas the earlier study by Nicks and O’Connell examined the impact of centrifugation on plating efficiency in noncompetitive cultures (5). It is possible centrifugation does not completely overcome the infectivity defect we detected in CTE202 but that the residual deficit is so small it can only be resolved with multiple passages. A growth defect in plasmid-deficient C. muridarum has been reported (50), so it is possible that a plasmid-associated growth defect exists in CTE202, although we could not detect differences in a synchronized developmental comparison.
We confirmed previous findings that Pgp4 is required for Pgp3 secretion (18). In the study by Lei et al., the pgp4 locus was replaced by pgp3, simultaneously placing pgp3 transcription under the control of the constitutive pgp4 promoter while deleting the Pgp4 coding sequence (18). However, pgp3 expression significantly increased at 22 h p.i.; thus, using inducible pgp3 in the presence and absence of pgp4 enabled us to recapitulate the normal expression profile of Pgp3 and exclude potential “off-target” host or chlamydial interactions that could influence the Pgp4 secretion phenotype.
The mechanism of Pgp3 secretion is still unclear. It has been speculated that Pgp3 is a potential type III secretion effector following observations of its secretion to the host cell cytosol (19). Other studies have suggested that some proteins encoded by plasmid-regulated chromosomal loci are also type III secretion effectors (51, 52), based upon their secretion by heterologous systems. However, a recent study proposed Pgp3 and other Pgp4-regulated proteins are secreted via OMVs in a Pgp4-dependent manner (18), following the identification of Pgp3 as a candidate OMV cargo protein (53, 54). OMVs are produced by Gram-negative bacteria to fulfill a variety of functions, including pathogenesis and host modulation (reviewed in reference 55). Although the precise mechanisms required for OMV production are still unclear, cargo loading into OMVs appears to be specific and coordinated. It is not known if or how Pgp4 directs this process, but it is possible that Pgp3 interacts directly with Pgp4 or with members of this secretion pathway whose expression is also Pgp4 dependent. Interaction studies could shed light on the mechanism of Pgp4-dependent secretion of Pgp3.
Our competition assays showed that loss of pgp3 expression results in an infectivity defect compared to wild type, an observation consistent with previously published studies (20, 21). Interestingly, uninduced CTE273, which does not express pgp3, was no more infectious than uninduced CTE277, which expresses neither pgp3 nor pgp4, suggesting expression of Pgp4 yields no infectious advantage in the absence of Pgp3. These results confirmed Pgp3 is required for plasmid-associated infectivity in C. trachomatis. Complementation via induction of pgp3 expression in CTE273 provided validation that induced Pgp3 functioned similarly to native Pgp3 in our studies.
Lack of pgp3 expression has been provided as an explanation for the infectivity defect observed in pgp4-deficient mutants (20). We expected that expression of Pgp3 alone would restore infectivity. Surprisingly, Pgp3 expression in the absence of Pgp4 did not overcome the infectivity defect in our in vitro competition assay. This suggested that Pgp3 is insufficient for plasmid-associated infectivity and indicated that the importance of Pgp4 to infectivity extends beyond regulation of pgp3. The design of our studies did not allow us to differentiate between a direct role for Pgp4 in infectivity or an indirect role via regulation of chromosomal loci. The functions of most Pgp4-regulated loci remain unknown, and their potential role in infectivity has not been explored. Therefore, it remains plausible that one or more of these loci could be involved in chlamydial infectivity, either independently of Pgp3 or within the same pathway. The latter scenario is supported by the need for both Pgp3 and Pgp4 to restore infectivity and our observation that a strain expressing Pgp4 alone is no more infectious than a Pgp3- and Pgp4-deficient strain. The proposed Pgp4-dependent OMV secretion pathway (18) connects Pgp3, Pgp4, and Pgp4-dependent chromosomal loci. A model in which optimal infectivity depends upon Pgp4-dependent secretion of Pgp3 is consistent with our observations.
The apparent importance for Pgp3 secretion to achieve wild-type levels of infectivity is puzzling. Previous studies suggested cytosolic Pgp3, which is released following cell lysis, binds to LL-37 (CRAMP in mice) in extracellular spaces and neutralizes its antichlamydial effects (11, 56). While this mechanism may contribute to the infectivity defect observed with pgp3-deficient strains in vivo, our studies used the murine fibroblast cell line L929, which does not secrete CRAMP (57). Therefore, Pgp3 binding to CRAMP following secretion cannot explain the infectivity defect observed in our assays. Other secreted chlamydial effectors with direct roles in known chlamydial infection pathways (28–34) are prepackaged in EBs and secreted via type III secretion following binding to the host cell. In contrast, Pgp3 is secreted late in the developmental cycle (19) and has not been identified as an early secreted effector in chlamydiae. Our experimental design also involved direct harvest of infected cells with mechanical lysis to avoid confounding effects that might arise from chlamydial plasmid-mediated EB release (24, 58). Isolation of plasmid-deficient strains of C. muridarum with suppressor mutations that restore wild-type infectivity further supports an indirect mechanism (6). Together, these data suggest that the role of Pgp3 in infectivity in our assays is likely indirect. However, the possibility that Pgp3 has a direct role in infectivity via an undiscovered role in the host cell cannot be disregarded based upon the results of this study or published observations.
Secretion of Pgp3 and other Pgp4-dependent secreted proteins coincides with RB-EB conversion. We reasoned that disruption of events at this crucial step in the developmental cycle could lead to the production of impaired EBs. The details of infectious EB production remain elusive, but based upon studies of other intracellular pathogens, we speculate that correct secretion and folding of an adhesin in the outer membrane could be important for optimally infectious EB production. It seems possible that disruption of OMV production, which requires intricate membrane dynamics, could lead to off-target effects, such as altered adhesin localization. We did not observe cytosolic OmcB in most of the cells infected with wild-type CM001 under any fixation condition. These results contrast with the observations published by Qi et al., demonstrating OmcB-specific detection in the cytosol of cells infected with wild-type C. trachomatis or C. muridarum (46). Slight variations between the staining protocol used in our study and the study by Qi et al. could lead to these differences, particularly since differences in fixation conditions led to increased cytosolic OmcB staining in CM972-infected cells. The increased cytosolic OmcB in CM972-infected cells with 2% PFA was likely an artifact of the fixation conditions. Similar observations have been made with CPAF, in which slow fixation at room temperature led to cytosolic CPAF but quick fixation did not (47). It is possible that increased cytosolic OmcB staining in CM972-infected cells fixed with 2% PFA, but not CM001- and CM3.1-infected cells, could reflect subtle changes in OmcB in CM972. While the artifactual nature of these observations hinders our ability to make conclusions about the role of OmcB in plasmid-associated infectivity, they do suggest future comparisons of membrane proteins in wild-type and plasmid-deficient chlamydiae should include OmcB. Alternatively, other adhesins, including polymorphic membrane proteins (59–61) and Ctad1 (62) could be altered in plasmid-deficient strains. Proteins involved in uptake and internalization or with indirect roles in adhesion could also serve as targets in our model.
Examination of changes to OmcB in the outer membrane and the role for other potential targets in our model would require a high-throughput screen and/or detailed studies of the EB membrane, both of which are beyond the scope of this study. If no differences in OmcB are observed between wild-type and plasmid-deficient chlamydiae, understanding the stage of the infection process that is altered in plasmid-, pgp3-, and pgp4-deficient chlamydiae (e.g., adhesion or internalization) and the point of secretion that is disrupted in pgp4-deficient chlamydiae could narrow the list of potential targets. Further, it would be interesting to test if deletion of genes encoding other Pgp4-dependent secreted proteins leads to an infectivity defect similar to that observed with pgp3 deletion. If this phenotype is conserved, it would provide additional evidence for an indirect role of OMV secretion in infectivity.
MATERIALS AND METHODS
Cell culture and bacterial strains.
Murine L929 fibroblasts, human HeLa cervical epithelial, and African green monkey Vero kidney cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 4.5 g/liter glucose, 4 mM l-glutamine, 110 mg/liter sodium pyruvate, and 10% fetal bovine serum.
All bacterial strains used in this study are summarized in Table 1. Chlamydiae were routinely propagated in cells passaged in 1× DMEM as described above unless otherwise noted. Infected cells were lysed by sonication, and bacteria were isolated by centrifugation. The resulting pellets were resuspended in Hanks’ balanced saline solution (HBSS) and treated with DNase1 and RNase for 1 h at 37°C before centrifugation at 20,000 × g for 20 min through a 32% Renografin cushion to remove residual cell membrane and other debris. After washing twice with HBSS, pellets were suspended in sucrose phosphate glutamic acid (SPG) buffer and stored at −80°C. Escherichia coli recombinants were cultured in Luria broth or on Luria agar containing 100 μg/mL ampicillin or 50 μg/mL spectinomycin, when appropriate.
TABLE 1.
Strains and plasmids used in this study
| Species | Strain | Plasmid | Genotype | Phenotypea | Source |
|---|---|---|---|---|---|
| Chlamydia trachomatis | CTE3024 | 67 | |||
| CTE3024 mCherry | p2TK2mCherry CmR | mCherry+, CmR | 67 | ||
| CTE3024 GFP | pGL2tetO | GFP+ ApR | This study, 26 | ||
| CTE202 | pCOC202 | CTE3024/Δ(pgp3-pgp4) | GFP+ ApR Pgp3− Pgp4− | This study | |
| CTE273 | pCOC273 | CTE3024/Ptet-pgp3 | mCherry+ SpR Pgp3+/− | This study | |
| CTE277 | pCOC277 | CTE3024/Ptet-pgp3 Δpgp4 | GFP+, ApR*, Pgp3+/−, Pgp4- | This study | |
| Chlamydia muridarum | CM001 | 68 | |||
| CM972 | Δ(pgp1-pgp8) | Plasmid deficient | 5 | ||
| CM3.1 | Δ(pgp1-pgp8) | Plasmid deficient | 6 | ||
| Escherichia coli | 10-β | New England Biolabs |
CmR, chloramphenicol resistant; ApR, ampicillin resistant; SpR, spectinomycin resistant; Pgp3+/−, expression was dependent upon the presence of aTc.
Construction of pCOC202, pCOC273, and pCOC277.
C. trachomatis serovar L2 harboring shuttle vector p2TK2Spec-SW2 mCh(Gro) Tet-Pgp3-3xF-IncDterm was a generous gift from Isabelle Derre (University of Virginia, Charlottesville, VA). pGL2tetO was kindly provided by Scott Hefty (University of Kansas, Lawrence, KS). To create pCOC202, the pgp3 and pgp4 loci were removed by PacI and PciI digestion and replaced with a random 100-bp nucleotide linker containing an AleI site using the oligonucleotides in Table S1. To create pCOC273 and pCOC277, DNA fragments were amplified from p2TK2Spec-SW2 mCh(Gro) Tet-Pgp3-3xF-IncDterm by PCR using the primers listed in Table S1, then assembled into the pGL2tetO vector using NEBuilder HiFi DNA assembly master mix (New England Biolabs, Ipswich, MA) according to manufacturer protocols. For shuttle vectors expressing SpR and mCherry, fragments containing the origin, aadA, and mCherry were amplified from p2TK2Spec-SW2 mCh(Gro) Tet-Pgp3-3xF-IncDterm and substituted for the region located between the PvuI and XhoI sites of pGL2tetO derivatives. Plasmid maps for pCOC202, pCOC273, and pCOC277 are provided in Fig. S1.
Transformation and clonal isolation of recombinant chlamydial strains by fluorescence-activated cell sorting.
The shuttle vectors pCOC202, pCOC273, pCOC277, and pGL2tetO were transformed into C. trachomatis CTE3024 as described by Cortina et al. (63). Primary transformants were selected with penicillin (10 U/mL) or spectinomycin (500 μg/mL)14 to 18 h p.i. After 40 h, cell monolayers were harvested and the bacteria were concentrated by centrifugation. Bacteria were infected onto Vero cell monolayers, incubated with antimicrobial selection, and centrifuged daily at 1,500 × g for 1 h at 37°C to optimize recovery and promote subsequent rounds of infection. Following harvest, transformants were sequentially passaged under antimicrobial selection to increase stock titers for clonal isolation.
Clonal populations representing each recombinant were derived by fluorescence-activated cell sorting (FACS) (64). Briefly, recombinants were infected into Vero cells at a low MOI and cultured with antibiotic selection for 24 to 28 h before flow cytometric sorting of fluorescent cells that contained the recombinant of interest expressing either GFP or mCherry, using a FACSAria III cell sorter (Becton, Dickinson, Franklin Lakes, NJ). Sorted cells were dispersed into individual wells of a 48-well plate containing a confluent monolayer of Vero cells at 1 cell per well and incubated at 37°C. Plates containing flow-sorted recombinants were cultured under antibiotic selection in medium containing 0.5 μg/mL cycloheximide and were centrifuged daily at 1,500 × g for 1 h at 37°C. Once expansion was detected within a well, the recombinants were harvested in SPG buffer and propagated to high titer by sequential passaging in L929 cells. FACS-isolated clones were validated by glycogen staining and by targeted sequencing of the Ptet promoter, pgp3, and pgp4 from recovered plasmids. Clone stocks were tested for the presence of Mycoplasma spp. by PCR using the Venor GeM mycoplasma detection kit (Sigma-Aldrich, St. Louis, MO).
Glycogen histochemistry.
C. trachomatis-infected cells were fixed at 40 h after infection and stained with iodine to detect intrainclusion glycogen accumulation (5). Inclusions were imaged using an Olympus BX53 fluorescence microscope.
Chlamydial quantitation.
Infectious progeny were quantified from growth curve samples, endocervical swabs, and cell culture samples as IFUs (65) in L929 monolayers by using a genus-specific monoclonal mouse anti-Chlamydia lipopolysaccharide clone, CF6J12 (1:500; Bio-Rad, Hercules, CA) and donkey anti-mouse IgG Alexa Fluor 488 (1:1,000; Life Technologies, Carlsbad, CA). To estimate total chlamydial abundance, including RB, nucleic acids were isolated from samples using a ZymoBiomics DNA/RNA Miniprep kit (Zymo Research, Irvine, CA). DNA was assayed by quantitative PCR using primers directed against omcA, and genome equivalents were estimated from a standard curve.
Quantitative RT-PCR.
Total RNA was extracted from C. trachomatis-infected cells using a ZymoBiomics DNA/RNA Miniprep kit with on-column DNase I treatment (Zymo) and converted to cDNA using an iScript gDNA clear cDNA synthesis kit (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. Gene expression from cDNA samples was assayed using primers for pgp3, pgp4, and 23S (Table S1) and advanced universal SYBR green supermix (Bio-Rad, Hercules, CA) in a Bio-Rad iCycler. Reactions were carried out under the following conditions: 95°C for 10 s and 55°C for 1 min, repeated for 40 cycles. Gene-specific accumulation of SYBR green-bound DNA was confirmed using melt curve analysis, and data were analyzed using Bio-Rad proprietary software.
Competition assay.
Competition assays were performed as previously described by Russell et al. (23), with some modifications. C. trachomatis strains mixed at a ratio of 1:10, 1:30, or 1:100 were infected into L929 monolayers at an MOI of ~2 with or without centrifugation at 1,500 × g for 60 min at 37°C, a condition that favors infection by plasmid-containing strains (5). Pgp3 expression was induced by adding 5 ng/mL aTc at 22 h p.i., when applicable. Cultures were incubated for 40 h at 37°C, then harvested in SPG buffer and placed at −80°C. After thawing, 1/10 or 1/100 of the passaged sample was used to infect new L929 monolayers, with or without centrifugation. A total of 5 sequential passages were performed for each experiment with two replicates per condition.
To quantify GFP- and mCherry-expressing C. trachomatis in each passage, aliquots were thawed and inoculated with centrifugation onto fresh L929 monolayers for direct visualization of chlamydial inclusions. GFP-expressing and mCherry-expressing inclusions were live imaged 24 to 26 h p.i. at 200× magnification using an Olympus CKX41 fluorescence microscope. A minimum of 3 fields were imaged to capture ≥50 inclusions for each sample. Green (GFP+) and red (mCherry+) inclusions were enumerated, and the abundance of GFP+ and/or mCherry+ inclusions present in each passage was expressed as a ratio. Fused GFP+ and mCherry+ inclusions, rarely observed in heavily infected monolayers, were excluded from analysis. To facilitate comparison between experiments, a competitive index (CI) for each competition assay was determined by dividing the ratio reflective of strain abundance generated at passage 5 by that obtained at passage 1.
Mouse model of genital tract infection.
Female C3H/HeJ mice were purchased from Jackson Laboratory (Bar Harbor, ME) and housed and cared for in accordance with experimental protocols approved by the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill. Age-matched 8- to 12-week-old mice were treated with 2.5 mg medroxyprogesterone (Amphastar Pharmaceuticals, Rancho Cucamonga, CA) by subcutaneous injection at least 7 days prior to infection to induce anestrous (66). Mice were infected transcervically (25) with 5× 106 IFU suspended in SPG using a nonsurgical embryo transfer device (Paratechs, Lexington, KY). Bacterial shedding from the lower genital tract was monitored by cervicovaginal swabbing over 10 days of infection.
Immunofluorescent staining of Pgp3 and OmcB.
To detect Pgp3 secretion into the cytosol, C. trachomatis-infected HeLa cells were fixed with 4% PFA at 37°C, 5% CO2 for 10 min. Cells permeabilized with 0.3% Triton X-100 were blocked overnight to reduce background staining and nonspecific binding. Proteins were detected with goat polyclonal anti-C. trachomatis MOMP antibody diluted 1:1,000 (Bio-Rad, Hercules, CA) and rabbit polyclonal anti-Pgp3 antibody (1:1,000) raised against purified recombinant protein. Specificity of the antisera was confirmed by immunoblotting of the recombinant proteins (Fig. S2A). Donkey anti-goat IgG and donkey anti-rabbit IgG secondary antibodies conjugated with Alexa Fluor 488 (Invitrogen, Waltham, MA) or Alexa Fluor 594 (Invitrogen, Waltham, MA) were used to label MOMP and Pgp3, respectively. For OmcB staining, HeLa cells were infected with C. muridarum strains at an MOI of 0.5 and fixed with 2% PFA for 1 h at room temperature or 4% PFA for 10 min at 37°C 5% CO2 at 40 h p.i. Fixed cells were permeabilized with 2% saponin for 1 h at room temperature. Blocked cells were stained with polyclonal mouse OmcB antiserum (1:3,000; kind gift from Ken Beagley, Queensland University of Technology, Brisbane, QLD, Australia) obtained from mice immunized with recombinant C-terminal OmcB (Lys212-Tyr554) (Fig. S2B and C) and Alexa Fluor 488-conjugated donkey anti-mouse IgG secondary antibodies (Invitrogen, Waltham, MA). Stained coverslips were mounted with ProLong glass antifade mountant with Nuc Blue (Invitrogen catalog number P36981) and imaged on an EVOS M7000 microscope.
Immunoblotting.
C. muridarum-infected L929 cells were treated with 10 μM lactacystin (Sigma-Aldrich, St. Louis, MO) 2 h prior to harvest in hot, fresh 1% SDS (with 150 mM NaCl, 50 mM Tris-HCl; pH 7.5) (48, 49). Protein concentrations of whole-cell lysates were measured using a Pierce bicinchoninic acid protein assay kit (Thermo Scientific, Waltham, MA). Equal amounts of protein were mixed with 4× lithium dodecyl sulfate loading buffer (Invitrogen, Waltham, MA) and dithiothreitol and heated at 95°C for 5 min before loading and separating on a 10% bis-Tris polyacrylamide gel by SDS-PAGE. Gel-resolved proteins were transferred to nitrocellulose membrane using a power blotter semidry transfer system (Thermo Scientific, Waltham, MA). Membranes were probed with polyclonal mouse OmcB antiserum (1:200), a species-specific monoclonal mouse anti-C. muridarum MOMP antibody clone M40 (1:500; kind gift from Elena Peterson [UC Irvine, CA]) (5), a monoclonal mouse anti-HSP60 antibody (1:1,000; Invitrogen, Waltham, MA), or a monoclonal rat anti-tubulin antibody clone MAB1864 (1:500; Millipore, Burlington, MA). Horseradish peroxidase-conjugated goat anti-mouse IgG (KindleBiosciences LLC, Greenwich, CT) and goat anti-rat IgG (Southern Biotech, Birmingham, AL) were used as secondary antibodies. Proteins of interest were visualized using the KwikQuant Western blot detection kit (Greenwich, CT) and the KwikQuant Imager (KindleBiosciences LLC, Greenwich, CT) according to the manufacturer’s instructions. Digital blue-on-black images were opened in Adobe Photoshop (San Jose, CA), converted to black-on-white images, and overlaid with corresponding white light images of the protein standards.
Statistical analyses.
All statistical analyses were conducted using GraphPad Prism 9 for Windows x64 version 9.3.1 (GraphPad Software, San Diego, CA). Competition assay analyses and growth rate determinations were accomplished using simple linear regression. Slopes calculated by linear regression were compared to determine differences between paired strains or conditions. Data from remaining experiments were analyzed by one- or two-way analysis of variance (ANOVA) followed by an appropriate test for multiple comparisons, unless otherwise noted. A P level of <0.05 represented a statistically significant difference between groups or samples regardless of the statistical test used.
ACKNOWLEDGMENTS
We thank Avinash Kollipara for assisting with FACS for clonal isolation. Sorting was completed at the UNC Flow Cytometry Core Facility, which is supported in part by P30 CA016086 Cancer Center Core support grant to the UNC Lineberger Comprehensive Cancer Center. We also thank our colleagues at the Queensland University of Technology, Logan Trim, Darren Leahy, Jonathan Harris, and Ken Beagley, for collecting and purifying the OmcB antiserum. The research described here was partially funded by a Boost award from the School of Medicine at the University of North Carolina at Chapel Hill. We thank Rob Nicholas for providing critical feedback on the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Catherine M. O’Connell, Email: catherine.oconnell@unc.edu.
Craig R. Roy, Yale University School of Medicine
REFERENCES
- 1.WHO. 2022. Trachoma. World Health Organization, Geneva, Switzerland. https://www.who.int/news-room/fact-sheets/detail/trachoma#:~:text=Trachoma%20is%20a%20disease%20of,of%20about%201.9%20million%20people. Accessed 1 July 2022. [Google Scholar]
- 2.CDC. 2022. Sexually transmitted disease surveillance 2020. U.S. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 3.Elwell C, Mirrashidi K, Engel J. 2016. Chlamydia cell biology and pathogenesis. Nat Rev Microbiol 14:385–400. 10.1038/nrmicro.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hybiske K, Stephens RS. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc Natl Acad Sci USA 104:11430–11435. 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Connell CM, Nicks KM. 2006. A plasmid-cured Chlamydia muridarum strain displays altered plaque morphology and reduced infectivity in cell culture. Microbiology (Reading) 152:1601–1607. 10.1099/mic.0.28658-0. [DOI] [PubMed] [Google Scholar]
- 6.O'Connell CM, Ingalls RR, Andrews CW, Scurlock AM, Darville T. 2007. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J Immunol 179:4027–4034. 10.4049/jimmunol.179.6.4027. [DOI] [PubMed] [Google Scholar]
- 7.Sigar IM, Schripsema JH, Wang Y, Clarke IN, Cutcliffe LT, Seth-Smith HMB, Thomson NR, Bjartling C, Unemo M, Persson K, Ramsey KH. 2014. Plasmid deficiency in urogenital isolates of Chlamydia trachomatis reduces infectivity and virulence in a mouse model. Pathog Dis 70:61–69. 10.1111/2049-632X.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei L, Chen J, Hou S, Ding Y, Yang Z, Zeng H, Baseman J, Zhong G. 2014. Reduced live organism recovery and lack of hydrosalpinx in mice infected with plasmid-free Chlamydia muridarum. Infect Immun 82:983–992. 10.1128/IAI.01543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Connell CM, Abdelrahman YM, Green E, Darville HK, Saira K, Smith B, Darville T, Scurlock AM, Meyer CR, Belland RJ. 2011. Toll-like receptor 2 activation by Chlamydia trachomatis is plasmid dependent, and plasmid-responsive chromosomal loci are coordinately regulated in response to glucose limitation by C. trachomatis. Infect Immun 79:1044–1056. 10.1128/IAI.01118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumoto A, Izutsu H, Miyashita N, Ohuchi M. 1998. Plaque formation by and plaque cloning of Chlamydia trachomatis biovar Trachoma. J Clin Microbiol 36:3013–3019. 10.1128/JCM.36.10.3013-3019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang C, Kari L, Lei L, Carlson JH, Ma L, Couch CE, Whitmire WM, Bock K, Moore I, Bonner C, McClarty G, Caldwell HD. 2020. Chlamydia trachomatis plasmid gene protein 3 is essential for the establishment of persistent infection and associated immunopathology. mBio 11. 10.1128/mBio.01902-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kari L, Whitmire WM, Olivares-Zavaleta N, Goheen MM, Taylor LD, Carlson JH, Sturdevant GL, Lu C, Bakios LE, Randall LB, Parnell MJ, Zhong G, Caldwell HD. 2011. A live-attenuated chlamydial vaccine protects against trachoma in nonhuman primates. J Exp Med 208:2217–2223. 10.1084/jem.20111266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira R, Borges V, Nunes A, Borrego MJ, Gomes JP. 2013. Assessment of the load and transcriptional dynamics of Chlamydia trachomatis plasmid according to strains’ tissue tropism. Microbiol Res 168:333–339. 10.1016/j.micres.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. 2010. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res 38:868–877. 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song L, Carlson JH, Whitmire WM, Kari L, Virtaneva K, Sturdevant DE, Watkins H, Zhou B, Sturdevant GL, Porcella SF, McClarty G, Caldwell HD. 2013. Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect Immun 81:636–644. 10.1128/IAI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Chen C, Gong S, Hou S, Qi M, Liu Q, Baseman J, Zhong G. 2014. Transformation of Chlamydia muridarum reveals a role for Pgp5 in suppression of plasmid-dependent gene expression. J Bacteriol 196:989–998. 10.1128/JB.01161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Zhang Q, Yang Z, Conrad T, Liu Y, Zhong G. 2015. Plasmid-encoded Pgp5 is a significant contributor to Chlamydia muridarum induction of hydrosalpinx. PLoS One 10:e0124840. 10.1371/journal.pone.0124840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei L, Yang C, Patton MJ, Smelkinson M, Dorward D, Ma L, Karanovic U, Firdous S, McClarty G, Caldwell HD, Ouellette SP. 2021. A chlamydial plasmid-dependent secretion system for the delivery of virulence factors to the host cytosol. mBio 12:e01179-21. 10.1128/mBio.01179-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Chen D, Zhong Y, Wang S, Zhong G. 2008. The chlamydial plasmid-encoded protein Pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect Immun 76:3415–3428. 10.1128/IAI.01377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Huang Y, Yang Z, Sun Y, Gong S, Hou S, Chen C, Li Z, Liu Q, Wu Y, Baseman J, Zhong G. 2014. Plasmid-encoded Pgp3 is a major virulence factor for Chlamydia muridarum to induce hydrosalpinx in mice. Infect Immun 82:5327–5335. 10.1128/IAI.02576-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsey KH, Schripsema JH, Smith BJ, Wang Y, Jham BC, O'Hagan KP, Thomson NR, Murthy AK, Skilton RJ, Chu P, Clarke IN. 2014. Plasmid CDS5 influences infectivity and virulence in a mouse model of Chlamydia trachomatis urogenital infection. Infect Immun 82:3341–3349. 10.1128/IAI.01795-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlson JH, Whitmire WM, Crane DD, Wicke L, Virtaneva K, Sturdevant DE, Kupko JJ, Porcella SF, Martinez-Orengo N, Heinzen RA, Kari L, Caldwell HD. 2008. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect Immun 76:2273–2283. 10.1128/IAI.00102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell M, Darville T, Chandra-Kuntal K, Smith B, Andrews CW, O'Connell CM. 2011. Infectivity acts as in vivo selection for maintenance of the chlamydial cryptic plasmid. Infect Immun 79:98–107. 10.1128/IAI.01105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang C, Starr T, Song L, Carlson JH, Sturdevant GL, Beare PA, Whitmire WM, Caldwell HD. 2015. Chlamydial lytic exit from host cells is plasmid regulated. mBio 6:e01648-15. 10.1128/mBio.01648-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gondek DC, Olive AJ, Stary G, Starnbach MN. 2012. CD4+ T cells are necessary and sufficient to confer protection against Chlamydia trachomatis infection in the murine upper genital tract. J Immunol 189:2441–2449. 10.4049/jimmunol.1103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickstrum J, Sammons LR, Restivo KN, Hefty PS. 2013. Conditional gene expression in Chlamydia trachomatis using the tet system. PLoS One 8:e76743. 10.1371/journal.pone.0076743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang C, Lei L, Collins JWM, Briones M, Ma L, Sturdevant GL, Su H, Kashyap AK, Dorward D, Bock KW, Moore IN, Bonner C, Chen C-Y, Martens CA, Ricklefs S, Yamamoto M, Takeda K, Iwakura Y, McClarty G, Caldwell HD. 2021. Chlamydia evasion of neutrophil host defense results in NLRP3 dependent myeloid-mediated sterile inflammation through the purinergic P2X7 receptor. Nat Commun 12. 10.1038/s41467-021-25749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y-S, Bastidas RJ, Saka HA, Carpenter VK, Richards KL, Plano GV, Valdivia RH. 2014. The Chlamydia trachomatis type III secretion chaperone Slc1 engages multiple early effectors, including TepP, a tyrosine-phosphorylated protein required for the pecruitment of CrkI-II to nascent inclusions and innate immune signaling. PLoS Pathog 10:e1003954. 10.1371/journal.ppat.1003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller KE, Fields KA. 2015. Application of β-lactamase reporter fusions as an indicator of effector protein secretion during infections with the obligate intracellular pathogen Chlamydia trachomatis. PLoS One 10:e0135295. 10.1371/journal.pone.0135295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci USA 101:10166–10171. 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh S, Ruelke EA, Ferrell JC, Bodero MD, Fields KA, Jewett TJ. 2020. Fluorescence-reported allelic exchange mutagenesis-mediated gene deletion indicates a requirement for Chlamydia trachomatis Tarp during in vivo infectivity and reveals a specific role for the C terminus during cellular invasion. Infect Immun 88:e00841-19. 10.1128/IAI.00841-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parrett CJ, Lenoci RV, Nguyen B, Russell L, Jewett TJ. 2016. Targeted disruption of Chlamydia trachomatis invasion by in trans expression of dominant negative Tarp effectors. Front Cell Infect Microbiol 6:84–84. 10.3389/fcimb.2016.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hower S, Wolf K, Fields KA. 2009. Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol Microbiol 72:1423–1437. 10.1111/j.1365-2958.2009.06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKuen MJ, Mueller KE, Bae YS, Fields KA. 2017. Fluorescence-reported allelic exchange mutagenesis reveals a role for Chlamydia trachomatis TmeA in invasion that is independent of host AHNAK. Infect Immun 85. 10.1128/IAI.00640-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su H, Raymond L, Rockey DD, Fischer E, Hackstadt T, Caldwell HD. 1996. A recombinant Chlamydia trachomatis major outer membrane protein binds to heparan sulfate receptors on epithelial cells. Proc Natl Acad Sci USA 93:11143–11148. 10.1073/pnas.93.20.11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su H, Watkins NG, Zhang YX, Caldwell HD. 1990. Chlamydia trachomatis-host cell interactions: role of the chlamydial major outer membrane protein as an adhesin. Infect Immun 58:1017–1025. 10.1128/iai.58.4.1017-1025.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su H, Zhang YX, Barrera O, Watkins NG, Caldwell HD. 1988. Differential effect of trypsin on infectivity of Chlamydia trachomatis: loss of infectivity requires cleavage of major outer membrane protein variable domains II and IV. Infect Immun 56:2094–2100. 10.1128/iai.56.8.2094-2100.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo C, Takahashi N, Swanson AF, Ozeki Y, Hakomori S. 1996. An N-linked high-mannose type oligosaccharide, expressed at the major outer membrane protein of Chlamydia trachomatis, mediates attachment and infectivity of the microorganism to HeLa cells. J Clin Invest 98:2813–2818. 10.1172/JCI119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanson AF, Kuo CC. 1994. Binding of the glycan of the major outer membrane protein of Chlamydia trachomatis to HeLa cells. Infect Immun 62:24–28. 10.1128/iai.62.1.24-28.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fadel S, Eley A. 2007. Chlamydia trachomatis OmcB protein is a surface-exposed glycosaminoglycan-dependent adhesin. J Med Microbiol 56:15–22. 10.1099/jmm.0.46801-0. [DOI] [PubMed] [Google Scholar]
- 41.Fechtner T, Stallmann S, Moelleken K, Meyer KL, Hegemann JH. 2013. Characterization of the interaction between the chlamydial adhesin OmcB and the human host cell. J Bacteriol 195:5323–5333. 10.1128/JB.00780-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang L, Liu D, Li Z, Zhou J, Tong D. 2021. Chlamydia abortus OmcB protein is essential for adhesion to host cells. J Basic Microbiol 61:1145–1152. 10.1002/jobm.202100312. [DOI] [PubMed] [Google Scholar]
- 43.Moelleken K, Hegemann JH. 2008. The Chlamydia outer membrane protein OmcB is required for adhesion and exhibits biovar-specific differences in glycosaminoglycan binding. Mol Microbiol 67:403–419. 10.1111/j.1365-2958.2007.06050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fechtner T, Galle JN, Hegemann JH. 2016. The novel chlamydial adhesin CPn0473 mediates the lipid raft-dependent uptake of Chlamydia pneumoniae. Cell Microbiol 18:1094–1105. 10.1111/cmi.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou S, Lei L, Yang Z, Qi M, Liu Q, Zhong G. 2013. Chlamydia trachomatis outer membrane complex protein B (OmcB) is processed by the protease CPAF. J Bacteriol 195:951–957. 10.1128/JB.02087-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi M, Gong S, Lei L, Liu Q, Zhong G. 2011. A Chlamydia trachomatis OmcB C-terminal fragment is released into the host cell cytoplasm and is immunogenic in humans. Infect Immun 79:2193–2203. 10.1128/IAI.00003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prusty BK, Chowdhury SR, Gulve N, Rudel T. 2018. Peptidase inhibitor 15 (PI15) regulates chlamydial CPAF activity. Front Cell Infect Microbiol 8:183. 10.3389/fcimb.2018.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson KA, Lee JK, Chen AL, Tan M, Sütterlin C. 2015. Induction and inhibition of CPAF activity during analysis of Chlamydia-infected cells. Pathog Dis 73:1–8. 10.1093/femspd/ftv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snavely EA, Kokes M, Dunn JD, Saka HA, Nguyen BD, Bastidas RJ, McCafferty DG, Valdivia RH. 2014. Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches. Pathog Dis 71:336–351. 10.1111/2049-632X.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skilton RJ, Wang Y, O'Neill C, Filardo S, Marsh P, Bénard A, Thomson NR, Ramsey KH, Clarke IN. 2018. The Chlamydia muridarum plasmid revisited: new insights into growth kinetics. Wellcome Open Res 3:25. 10.12688/wellcomeopenres.13905.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Da Cunha M, Milho C, Almeida F, Pais SV, Borges V, Maurício R, Borrego MJ, Gomes JP, Mota LJ. 2014. Identification of type III secretion substrates of Chlamydia trachomatis using Yersinia enterocolitica as a heterologous system. BMC Microbiol 14:40. 10.1186/1471-2180-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gehre L, Gorgette O, Perrinet S, Prevost M-C, Ducatez M, Giebel AM, Nelson DE, Ball SG, Subtil A. 2016. Sequestration of host metabolism by an intracellular pathogen. Elife 5. 10.7554/eLife.12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frohlich KM, Hua Z, Quayle AJ, Wang J, Lewis ME, Chou CW, Luo M, Buckner LR, Shen L. 2014. Membrane vesicle production by Chlamydia trachomatis as an adaptive response. Front Cell Infect Microbiol 4:73. 10.3389/fcimb.2014.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frohlich K, Hua Z, Wang J, Shen L. 2012. Isolation of Chlamydia trachomatis and membrane vesicles derived from host and bacteria. J Microbiol Methods 91:222–230. 10.1016/j.mimet.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hou S, Dong X, Yang Z, Li Z, Liu Q, Zhong G. 2015. Chlamydial plasmid-encoded virulence factor Pgp3 neutralizes the antichlamydial activity of human cathelicidin LL-37. Infect Immun 83:4701–4709. 10.1128/IAI.00746-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heap RE, Marín-Rubio JL, Peltier J, Heunis T, Dannoura A, Moore A, Trost M. 2021. Proteomics characterisation of the L929 cell supernatant and its role in BMDM differentiation. Life Sci Alliance 4:e202000957. 10.26508/lsa.202000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pereira IS, Pais SV, Borges V, Borrego MJ, Gomes JP, Mota LJ. 2022. The type III secretion effector CteG Mediates host cell lytic exit of Chlamydia trachomatis. Front Cell Infect Microbiol 12:902210. 10.3389/fcimb.2022.902210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mölleken K, Schmidt E, Hegemann JH. 2010. Members of the Pmp protein family of Chlamydia pneumoniae mediate adhesion to human cells via short repetitive peptide motifs. Mol Microbiol 78:1004–1017. 10.1111/j.1365-2958.2010.07386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mölleken K, Becker E, Hegemann JH. 2013. The Chlamydia pneumoniae invasin protein Pmp21 recruits the EGF receptor for host cell entry. PLoS Pathog 9:e1003325. 10.1371/journal.ppat.1003325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Becker E, Hegemann JH. 2014. All subtypes of the Pmp adhesin family are implicated in chlamydial virulence and show species-specific function. Microbiologyopen 3:544–556. 10.1002/mbo3.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stallmann S, Hegemann JH. 2016. The Chlamydia trachomatis Ctad1 invasin exploits the human integrin β1 receptor for host cell entry. Cell Microbiol 18:761–775. 10.1111/cmi.12549. [DOI] [PubMed] [Google Scholar]
- 63.Cortina ME, Ende RJ, Bishop RC, Bayne C, Derré I. 2019. Chlamydia trachomatis and Chlamydia muridarum spectinomycin resistant vectors and a transcriptional fluorescent reporter to monitor conversion from replicative to infectious bacteria. PLoS One 14:e0217753. 10.1371/journal.pone.0217753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alzhanov DT, Suchland RJ, Bakke AC, Stamm WE, Rockey DD. 2007. Clonal isolation of Chlamydia-infected cells using flow cytometry. J Microbiol Methods 68:201–208. 10.1016/j.mimet.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 65.Kelly KA, Robinson EA, Rank RG. 1996. Initial route of antigen administration alters the T-cell cytokine profile produced in response to the mouse pneumonitis biovar of Chlamydia trachomatis following genital infection. Infect Immun 64:4976–4983. 10.1128/iai.64.12.4976-4983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tuffrey M, Taylor-Robinson D. 1981. Progesterone as a key factor in the development of a mouse model for genital-tract infection with Chlamydia trachomatis. FEMS Microbiol Lett 12:111–115. 10.1111/j.1574-6968.1981.tb07622.x. [DOI] [Google Scholar]
- 67.McQueen BE, Kiatthanapaiboon A, Fulcher ML, Lam M, Patton K, Powell E, Kollipara A, Madden V, Suchland RJ, Wyrick P, O'Connell CM, Reidel B, Kesimer M, Randell SH, Darville T, Nagarajan UM. 2020. Human Fallopian tube epithelial cell culture model to study host responses to Chlamydia trachomatis infection. Infect Immun 88. 10.1128/IAI.00105-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sullivan J, Frazer L, Darville T, O'Connell C. 2014. Chlamydia muridarum used in the mouse model of C. trachomatis, contains multiple variants that lead to differential immune responses and disease development. J Immunol 192:67.6–67.6. 10.4049/jimmunol.192.Supp.67.6. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 and Fig. S1 to S5. Download iai.00392-22-s0001.pdf, PDF file, 5.5 MB (5.7MB, pdf)