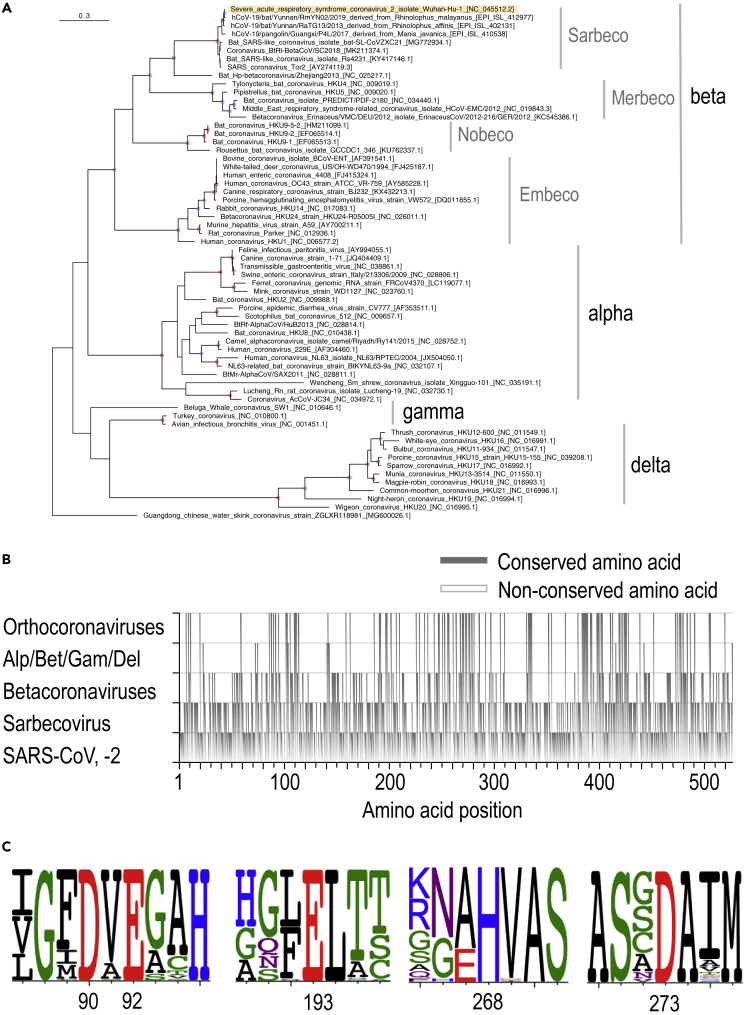
Figure 1.
Phylogenetic tree and amino acid sequence alignment of nsp14 derived from representative coronaviruses
(A) Maximum likelihood (ML)-based phylogenetic tree of the 62 representative coronaviruses. Amino acid sequences of nsp14 in ORF1ab were used for this analysis. A red or blue circle in an internal node corresponds to bootstrap values ≥95% or ≥80%, respectively. The virus located at the top is SARS-CoV-2 (hCoV-19/Wuhan/Hu-1/2019).
(B) Conserved amino acid residues among each group. The amino acid sequence of the nsp14 of the 62 representative coronaviruses is shown according to the relative amino acid positions of SARS-CoV-2 nsp14 (note that the gaps in the SARS-CoV-2 sequence are not shown). The conserved amino acid positions are indicated in gray.
(C) Five conserved active site residues involved in proofreading. The amino acid proportions for each site were calculated based on the sequence alignment of the nsp14 of the 62 representative coronaviruses visualized by using WebLogo.14 The amino acids are colored according to their chemical properties. See also Figure S2 for the entire alignment.