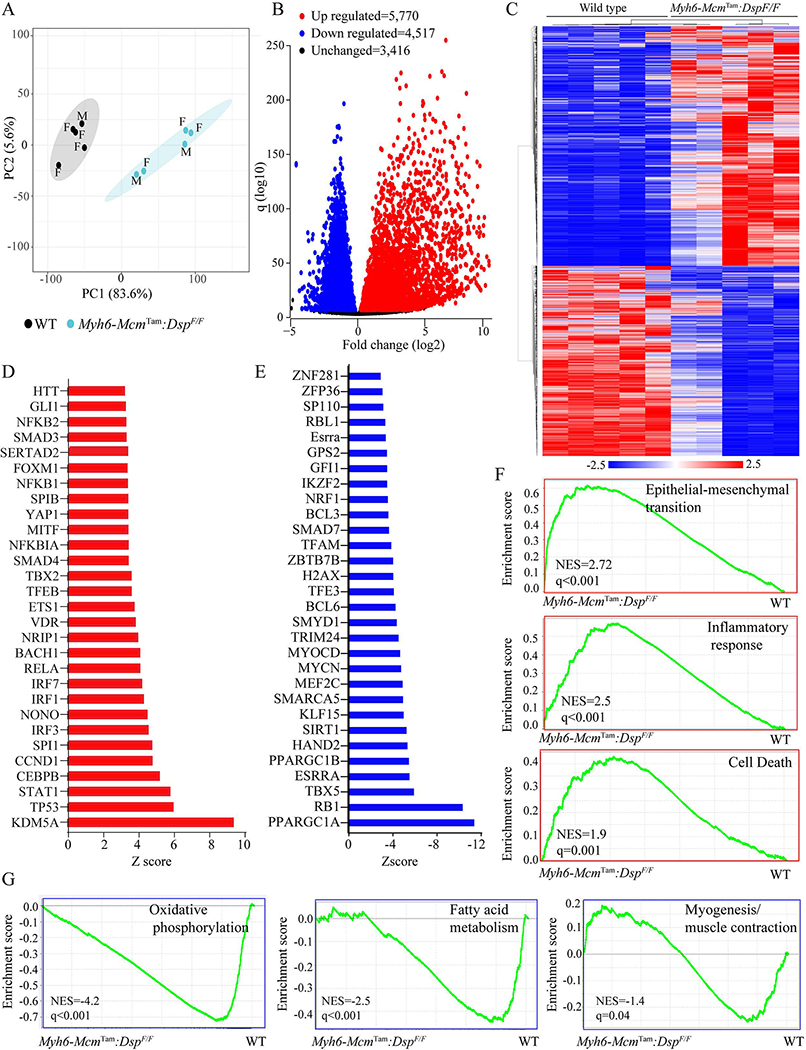
Figure 2.
Cardiac myocyte transcripts analyzed by RNA sequencing. (A) Principal Component Analysis (PCA) of the transcripts of cardiac myocytes isolated from 4-week-old WT and Myh6-McmTam:DspF/F mice. Male (M) and Female (F) mice are marked. (B) Volcano plot of the differentially expressed genes (DEGs) between the WT and Myh6-McmTam:DspF/F myocytes. The upregulated genes are shown in red, the down-regulated ones in blue, and the unchanged ones in black. (C) Heat map of the DEGs between the WT and Myh6-McmTam:DspF/F myocytes. The scale is shown at the bottom of the map. (D) List of the transcriptional regulators of genes expression predicted to be activated based on the DEGs in the Myh6-McmTam:DspF/F myocytes. (E) List of the transcriptional regulators of genes expression predicted to be suppressed based on the DEGs in the Myh6-McmTam:DspF/F myocytes. (F) Gene set enrichment analysis (GSEA) maps showing enrichment of genes involved in the epithelial-mesenchymal transition (EMT), inflammatory response, and cell death in the Myh6-McmTam:DspF/F myocytes. (G) GSEA maps predicting suppression of oxidative phosphorylation, fatty acid metabolism, and myogenesis (muscle contraction) in the Myh6-McmTam:DspF/F myocytes.