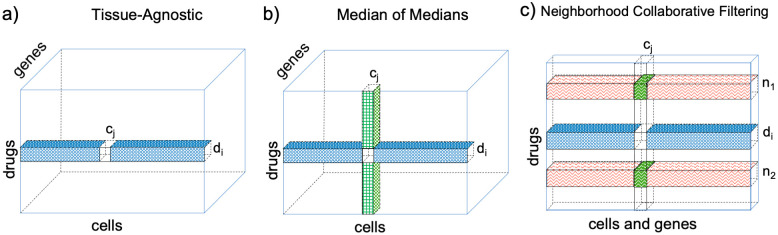
Fig 1. Methods overview.
a) In Tissue-Agnostic imputation, the median of the expression of the first gene for drug di in cells other than cj (the blue-colored row in the 3D matrix M) is used to predict the unknown expression profile for that cell/drug/gene combination (shown as a white missing value in the blue row). b) The Median of Medians (MoM) method predicts the same missing value by taking the median of two values: the median of that gene’s expression for drug di in cells other than cj (the blue-colored row) and the median of that gene’s expression for drugs other than di in cell cj (the green-checked column). c) In Neighborhood Collaborative Filtering, we use the flattened 2D matrix R, where rows contain values for all genes in cell 1, all genes in cell 2, and so on. Neighbors are drugs whose expression profiles on cells other than cj (orange waves) are most similar to drug di’s profile on cells other than cj (the blue-colored row). In this example with k = 2 neighbors, the expression profiles for cell cj on neighbor drugs dn1 and dn2 (darker / green waves) are used to impute the missing profile for drug di on cell cj.