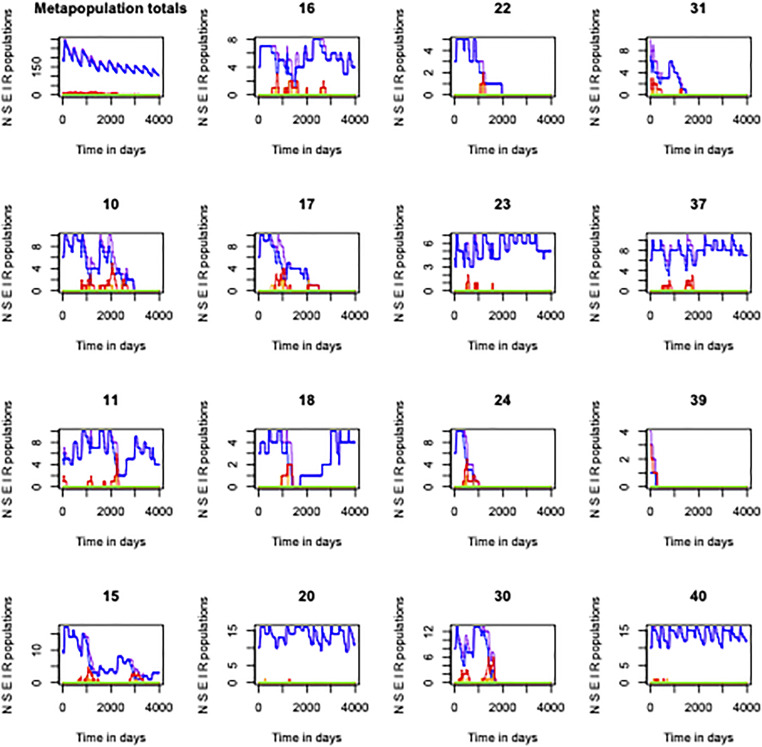
Fig 5. Examples of runs of simulation metaseir, which is called by seirshell.
The figure plots overall metapopulation numbers and outcomes in 15 patches (excluding patches where local K = 0 and including diversity among the remaining patches with respect to K) to illustrate the types of results that can be obtained. N, S, E, I, R numbers are shown in purple, blue, orange, red, green respectively. This simulation run with β = 0.0015, ab = 0.6, m = 0.00005 and α = 0.001 led to an epidemic persistence time of 3350 days (9.2 years), and the kit fox population dropped to about half of its starting point. Both kit fox and parasite populations show metapopulation dynamics with local extinction and colonization, with the annual birth pulse evident in the metapopulation totals panel. Data were generated in R by the function call: > h = 49; fox49outmarch2 = seirshell(trials = 1,tmax = 4000,h = 49, beta = 0.0015, alphabeta = 0.6, EIbirth = F,BetaHwander = diag(0,h,h), seirscape = foxscape49, seirdem = foxseirdem49, seirrates = foxseirrates49, seirnaught = foxseirnaught49); animetaseir(fox49outmarch2$meanmfx,4,4, patchdisplay = c(10,11,15,16,17,18,20,22,23,24,30,31,37,39,40),deathtrace = F);fox49outmarch2$epitimes.