Figure 3: Identification of functional regulatory variants with reproducible and concordant effects in allele-specific stimulation of transcriptional activity.
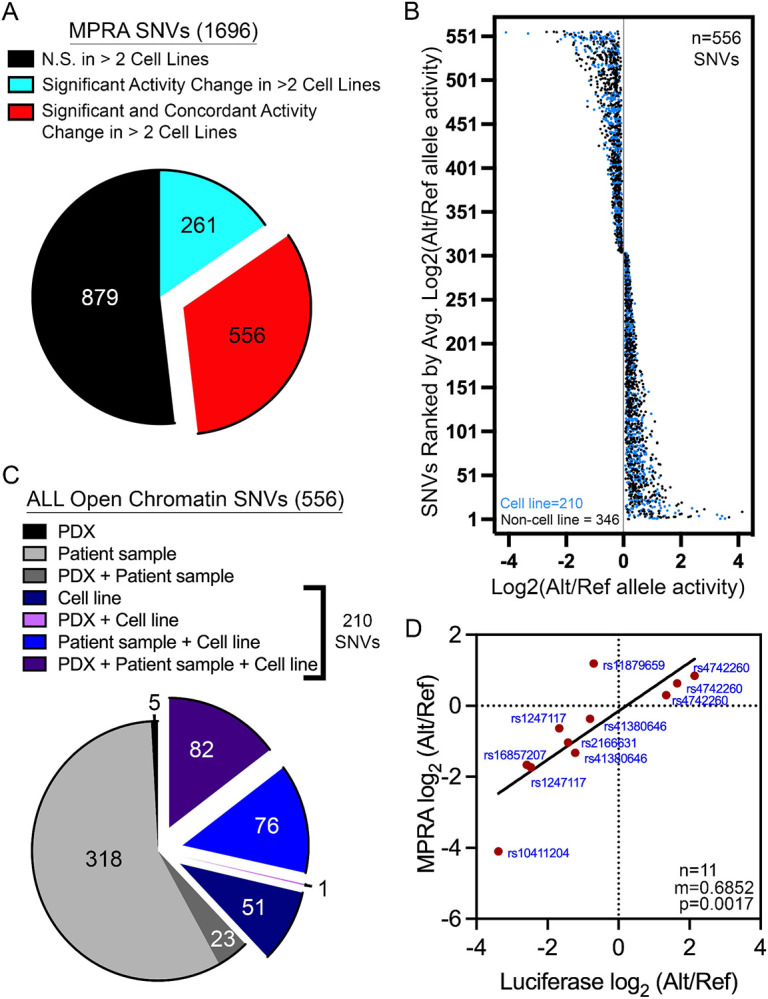
(A) 556 of the 1696 SNVs assayed are functional regulatory variants with reproducible (FDR<0.05 in >2 cell lines) and concordant (same directionality in >2 cell lines) changes in allele-specific activity. (B) Plot showing the distribution of log2-adjusted activity between alternative (Alt) and reference (Ref) alleles across 556 functional regulatory variants. (C) Pie chart shows how many functional regulatory variants map to open chromatin in diverse ALL cell models. 210 of the 556 functional regulatory variants are found in accessible chromatin sites that was identified in an ALL cell line. (D) Top hits from the 210 functional regulatory variants found in accessible chromatin in ALL cell lines were orthogonally validated by luciferase reporter assays. Data show significant correlation between the allele-specific effects detected by MPRA and dual-luciferase reporter assays.
