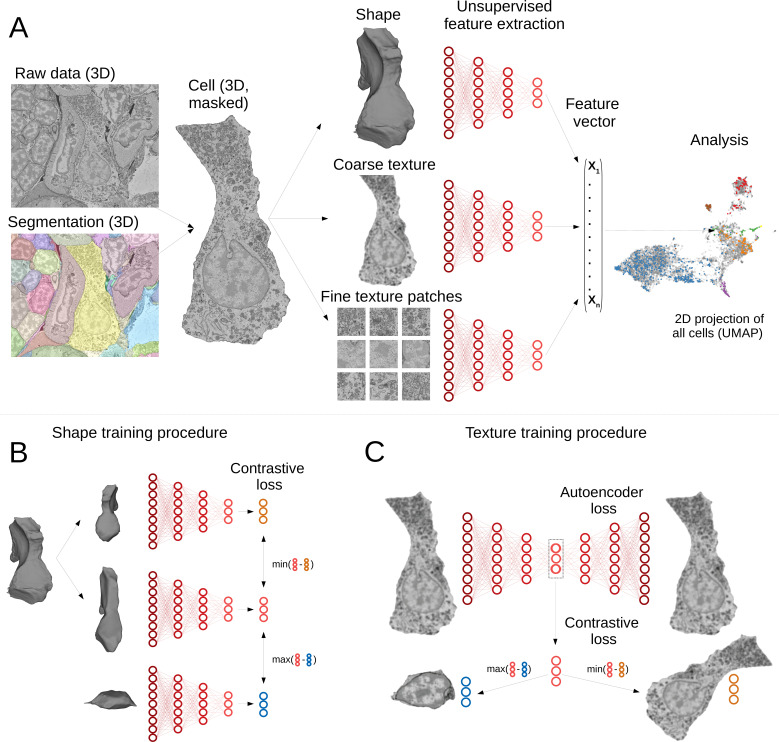
Figure 1. Deep-learning pipeline for extracting MorphoFeatures.
(A) Cell segmentation is used to mask the volume of a specific cell (and its nucleus) in the raw data. Neural networks are trained to represent shape, coarse, and fine texture from the cell volume (separately for cytoplasm and nuclei). The resulting features are combined in one MorphoFeatures vector that is used for the subsequent analysis. (B) Training procedure for the shape features. A contrastive loss is used to decrease the distance between the feature vectors of two augmented views of the same cell and increase the distance to another cell. (C) Training procedure for the coarse and fine texture features (here illustrated by coarse texture). Besides the contrastive loss, an autoencoder loss is used that drives the network to reconstruct the original cell from the feature vector.