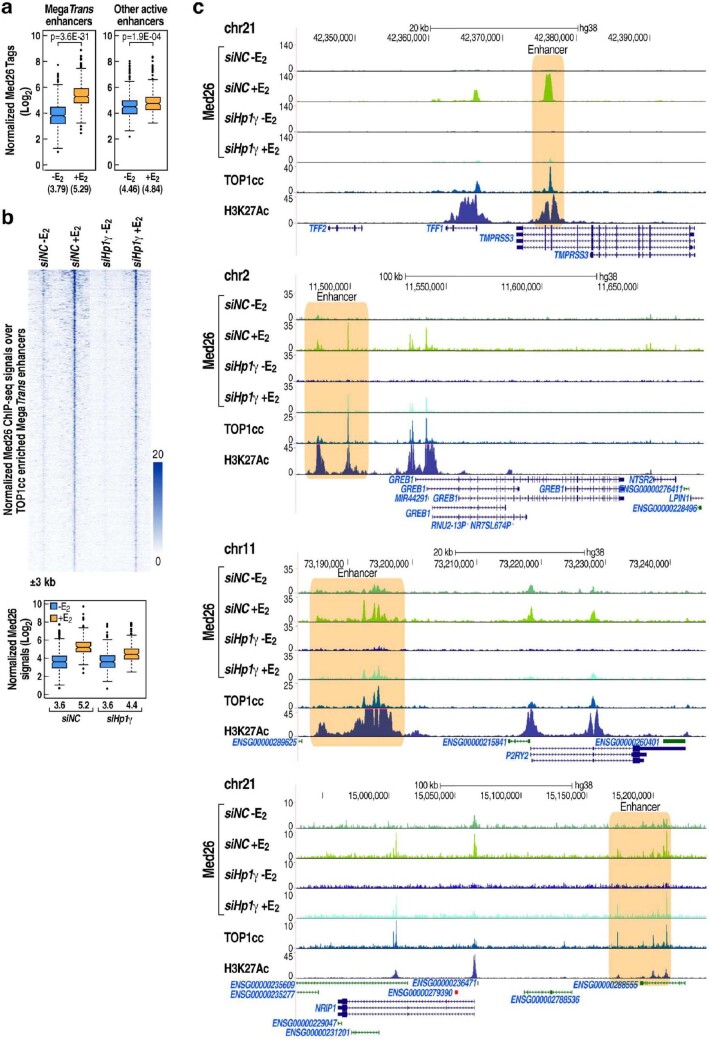
Extended Data Fig. 8. HP1γ is required for the tethering of Med26 to the acutely activated enhancers.
(a) Box-and-whisker plots show Med26 ChIP-seq signals at and TOP1cc-enriched MegaTrans enhancers (n = 481) and other active enhancers (n = 3,533), which have similar eRNA expression levels with E2 treated TOP1cc-enriched MegaTrans enhancers in MCF7 cells. P values generated from unpaired two-tailed t test denote statistical differences between −E2 and +E2 conditions in the box-and-whisker plots. The median value of normalized Med26 ChIP-seq tags (Log2) are listed under the boxplots. (b) Heatmaps and box-and-whisker plots show Med26 ChIP-seq signals at TOP1cc-enriched MegaTrans enhancers (n = 481). Additional 3 kb from the center of the peaks are shown, and the color scale shows the normalized tag numbers, P values generated from unpaired two-tailed t test denote statistical differences between shNC + E2 and siHp1γ + E2 conditions in the box-and-whisker plots. The median values of normalized Med26 ChIP-seq tags (Log2) are listed under the boxplots. (c) Genomic browsers show Med26 ChIP-seq, H3K27Ac ChIP-seq, and TOP1cc CUT&RUN signals at the selected gene locus. Enhancers are highlighted with light-brown box. For a and b, center lines show the medians, box limits indicate the 25th and 75th percentiles, and whiskers extend 1.5× the interquartile range from the 25th and 75th percentiles. The median values of normalized Med26 ChIP-seq tags (Log2) are listed under the boxplots.