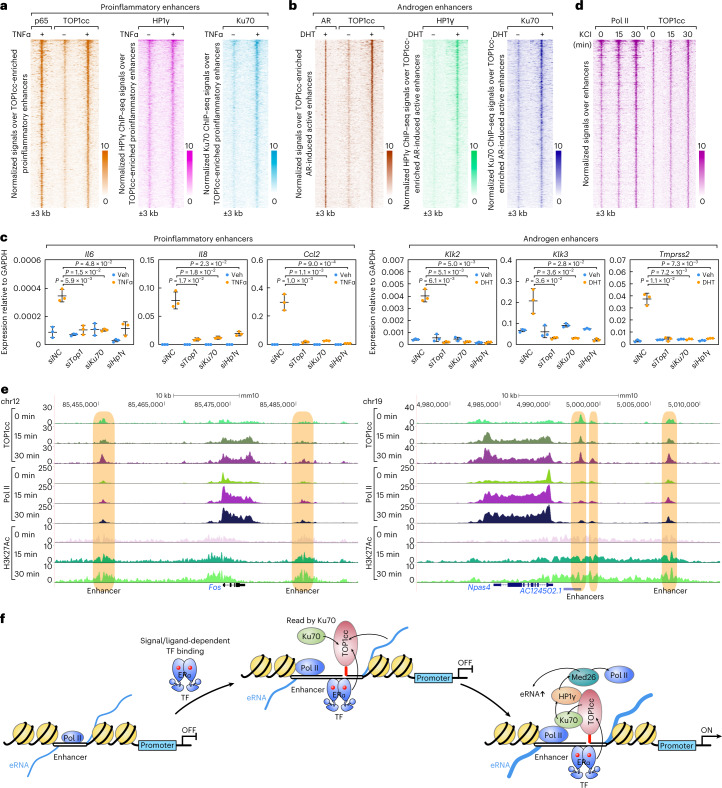
Fig. 5. TOP1cc–Ku70–HP1γ serves as a general transcriptional code underlying acute transcriptional activation.
a, Heatmaps show TOP1cc CUT&RUN signals and ChIP–seq signals (p65, HP1γ and Ku70) at TOP1cc-enriched acute TNFα activated proinflammatory enhancers (n = 589). b, Heatmaps show TOP1cc CUT&RUN signals and ChIP–seq signals (AR, HP1γ and Ku70) at TOP1cc-enriched acute DHT activated androgen enhancers (n = 691). An additional 3 kb from the center of the peaks are shown. c, RT–qPCR results show that the TNFα induced Il6, Il8 and Ccl2 genes, and DHT-induced Klk2, Klk3 and Tmprss2 genes are impaired by knockdown of Top1, Ku70 or Hp1γ. Data are shown as mean ± s.d. (n = 3 (three independent biological replicates), two-tailed Student’s t-test). d, Heatmaps show TOP1cc CUT&RUN signals and Pol II ChIP–seq signals at TOP1cc-enriched KCl-induced active neuronal enhancers (n = 731). e, Genome browser images show Top1 CUT&RUN signals and ChIP–seq signals (Pol II and H3K27Ac) at the selected neuronal gene loci upon acute KCl treatment. Enhancers are highlighted with light-brown boxes. f, Working model: the signal-dependent acutely activated enhancers requires the signal-dependent transcriptional factor and enhancer RNA-dependent formation of TOP1cc, functioning as a broadly required transcriptional code read by Ku70, enabling the recruitment of HP1γ–Med26 for signal-dependent transcriptional activation. For a, b and d, an additional 3 kb from the center of the peaks are shown in the heatmaps, and the color scale shows the normalized tag numbers. Raw data for graphs in c are available as Source data.