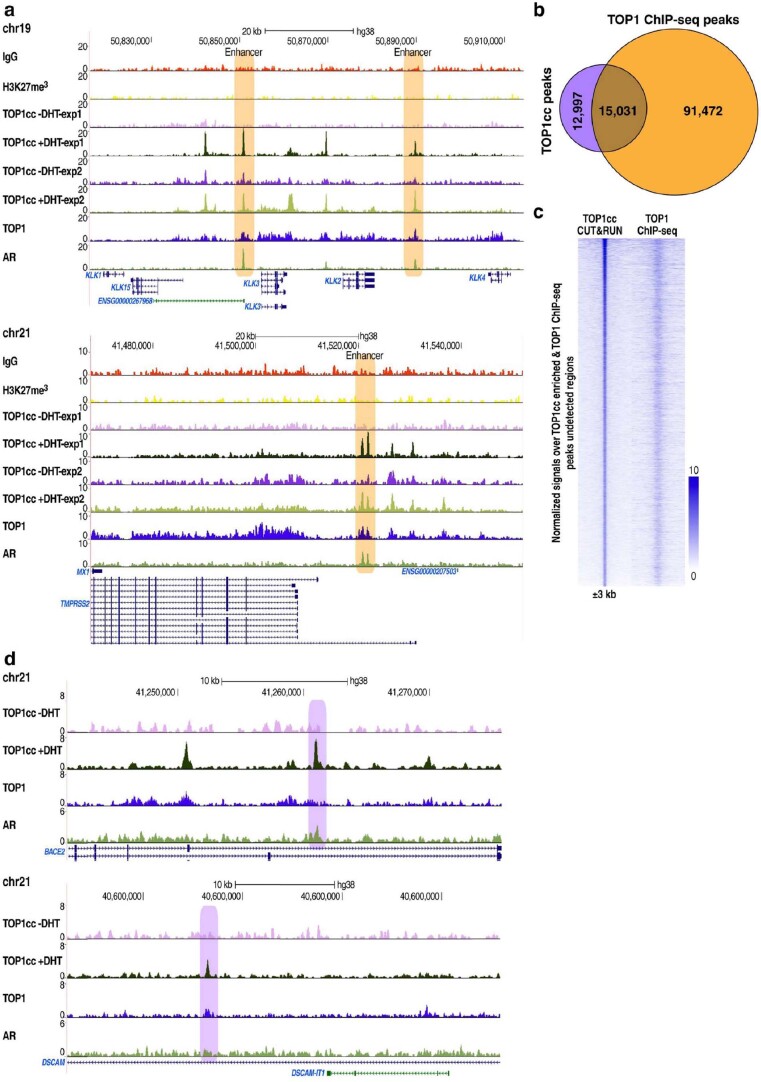
Extended Data Fig. 1. CUT&RUN assays disclose TOP1cc signals in LNCAP cells.
(a) Genomic browsers show TOP1cc CUT&RUN signals, AR and Top1 ChIP-seq signals in DHT treated LNCAP cells. IgG and H3K27me3 CUT&RUN signals are serving as the experimental controls for the CUT&RUN assays. Enhancers are highlighted with light-brown boxes. (b) Co-localization analysis of Top1 ChIP-seq signals and TOP1cc CUT&RUN signals in DHT treated LNCAP cells. (c) Heatmaps show Top1 ChIP-seq signals and TOP1cc CUT&RUN signals at TOP1cc-enriched but Top1 ChIP-seq peak undetected regions. Additional 3 kb from the center of the peaks are shown, and the color scale shows the normalized tag numbers. (d) Genomic browsers show TOP1cc CUT&RUN signals, AR and Top1 ChIP-seq signals in DHT treated LNCAP cells. TOP1cc-enriched but Top1 peaks undetected regions are highlighted with light-purple boxes.