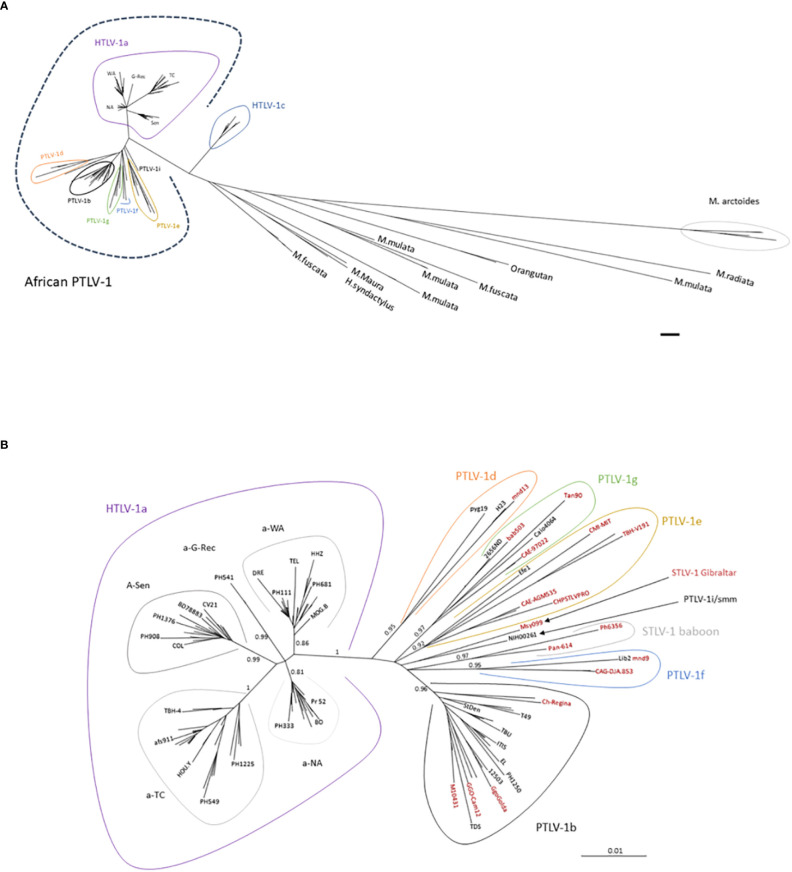
Figure 9.
(A) General disposition of African PTLV-1 among the other PTLV-1. Phylogenetic tree generated from a 711-nucleotide long alignment of 196 sequences. Sequences comprise African PTLV-1, Australo-Melanesian HTLV-1c and Asian Macaque STLV-1 strains. The phylogenetic tree was generated using the Maximum Likelihood method. The scale bar represents nucleotide substitutions per site. (B) Phylogenetic tree presenting the genetic diversity of African PTLV-1. The analysis was performed on an alignment of 125 HTLV-1 and 30 STLV-1 sequences (presented in black and red respectively). This alignment contains distinct African HTLV-1 full-length LTR sequences (in black), and the most representative STLV-1 full-length sequences (in red). The size of the complete LTR available in GenBank varies between 745-nt and 790-nt depending on the strain, resulting in an alignment of 772-nt nucleotides (due to difficulties in aligning some regions). The phylogenetic tree was derived by the neighbor-joining method using the GTR model (gamma = 0.5017), with the PAUP software. Horizontal branch lengths are drawn to scale, which is indicated by the bar. The topology of the phylogenetic tree was validated by Maximum Likelihood (using the SeaView5 software), and the strength of the branches was estimated using approximate likelihood-ratio test (aLRT). The values correspond to the calculated probability. The definition of genotypes (or subtypes of PTLV-1) depends on the number of sequences present for each group, and the size of the sequences. The historically coined “South African and East African STLV-1” subgroup (219) is now presented as part of the PTLV-1e genotype. Similarly, the “Central and West Africa STLV-1” subgroups now belong to the HTLV-1g genotype. All genotypes represented in Africa are presented in the tree, except HTLV-1h (as described by Liégeois (220), for which only partial LTR sequences are available. HTLV-1a is strictly present in humans, STLV-1 Gibraltar and STLV-1 baboon have never been detected in humans. All the other subgroups have been identified both in human and simian hosts.